Abstract
Sequential events in macromolecular biosynthesis are often elegantly coordinated. The small ribosomal subunit (SSU) processome is a large ribonucleoprotein (RNP) required for processing of precursors to the small subunit RNA, the 18S, of the ribosome. We have found that a subcomplex of SSU processome proteins, the t-Utps, is also required for optimal rRNA transcription in vivo in the yeast Saccharomyces cerevisiae. The t-Utps are ribosomal chromatin (r-chromatin)-associated, and they exist in a complex in the absence of the U3 snoRNA. Transcription is required neither for the formation of the subcomplex nor for its r-chromatin association. The t-Utps are associated with the pre-18S rRNAs independent of the presence of the U3 snoRNA. This association may thus represent an early step in the formation of the SSU processome. Our results indicate that rRNA transcription and pre-rRNA processing are coordinated via specific components of the SSU processome.
Keywords: Ribosome, RNA, transcription, RNA processing, SSU processome
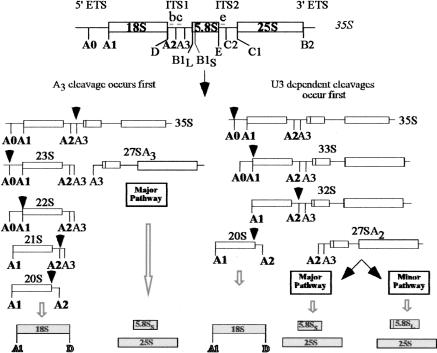
The overall scheme of ribosome biogenesis is largely conserved from the most primitive of eukaryotes to mammals. It requires the orchestrated steps of transcription, endonucleolytic and exonucleolytic cleavages, rRNA methylations and pseudouridylations, and folding of the ribosomal RNA (for review, see Gerbi et al. 2001). In single-celled organisms such as the yeast Saccharomyces cerevisiae, a great deal of energy is expended to generate the components of ribosomes (Warner 1999,2001). Processing of the nascent rRNA (35S) into the mature 18S, 5.8S, and 25S rRNAs of the ribosome is shown in Figure 1 (for review, see Kressler et al. 1999; Venema and Tollervey 1999). The steps in the pre-rRNA processing pathway are not temporally obligatory and vary somewhat according to strain background. As indicated in Figure 1, either the U3-dependent cleavages at A0, A1, and A2 or cleavage at A3 in ITS 1 can occur first (Dunbar et al. 1997; Lee and Baserga 1997). In the latter case, the 23S rRNA precursor and the 27SA3 pre-rRNAs are generated. The 23S rRNA is a precursor to the 18S rRNA of the small ribosomal subunit (SSU), and the 27SA rRNAs are precursors to the 5.8S and 25S rRNAs of the large ribosomal subunit (LSU). Following the separating cleavage either at A2 or A3, the two pre-rRNAs, representing two sides of the processing pathway, are then processed independently. The 20S pre-rRNA is exported to the cytoplasm and processed there into the 18S rRNA. The 18S rRNA and its associated ribosomal proteins constitute the SSU of the yeast ribosome (40S). The rRNA precursors to the 5.8S and 25S rRNAs undergo a number of processing steps, and together with their associated proteins and the 5S rRNA, are assembled into the LSU of the yeast ribosome (60S).
Figure 1.
Pre-rRNA processing in S. cerevisiae. In yeast the rDNA is found in a 9.1-kb unit repeated 100–200 times. The 35S pre-rRNA is transcribed by RNA polymerase I (Pol I). In some instances the 35S is cleaved at A3, yielding the 23S pre-rRNA, which is then processed at A0, A1, and A2 to yield the 20S precursor. In other instances, the 35S pre-rRNA is processed sequentially at A0, A1, and A2 to yield the 20S precursor. The 20S precursor is exported to the cytoplasm and matured into the 18S rRNA as part of the small ribosomal subunit (SSU). The 27SA3 pre-rRNA is then processed to generate the 5.8S and 25S rRNAs of the large ribosomal subunit (LSU). The 27SA2 precursor is processed into the 5.8S and 25S rRNAs of the LSU. The U3 snoRNA-dependent cleavages are in bold. The location of oligonucleotides b, c, and e used to probe for pre-rRNAs on Northern blots is indicated.
There are over 100 proteins required for processing of the 35S pre-rRNA (for review, see Granneman and Baserga 2004). The complexes required for the processing of these precursors have recently been purified by several laboratories, all of which have exploited epitope tagging and mass spectrometry techniques (Bassler et al. 2001; Harnpicharnchai et al. 2001; Dragon et al. 2002; Fatica et al. 2002; Gavin et al. 2002; Grandi et al. 2002; Ho et al. 2002; Nissan et al. 2002; Wu et al. 2002; Milkereit et al. 2003a,b; Peng et al. 2003; Saveanu et al. 2003; Schafer et al. 2003; Tschochner and Hurt 2003).
The SSU processome is a large ribonucleoprotein (RNP) containing the U3 snoRNA and at least 40 proteins or Utps (U three protein) required for 40S ribosomal subunit biogenesis (Dragon et al. 2002; K. Bernstein, J. Gallagher, and S. Baserga, unpubl.). A similar RNP has also been purified and referred to as the 90S pre-ribosome (Gavin et al. 2002). The U3 snoRNA is a noncanonical box C/D snoRNA, conserved from trypanosomes to humans, essential for the pre-rRNA cleavages that produce the 18S rRNA (A0, A1, and A2 in yeast; Hughes and Ares 1991). The U3 snoRNA base pairs directly with prerRNA at two discrete sites (Beltrame and Tollervey 1992; Beltrame et al. 1994; Hughes 1996; Sharma and Tollervey 1999). Two different-sized complexes harboring the U3 snoRNA can be detected in cell extracts: a small nucleolar RNP (12S; referred to as the U3 snoRNP or mono-snoRNP; Watkins et al. 2000) and a large RNP, the SSU processome (80S; Dragon et al. 2002). The large cohort of proteins complexed with the U3 snoRNA is essential for SSU processome function in the nucleolus (for review, see Culver 2002; Peculis 2002). The SSU processome can be visualized in chromatin (Miller) spreads as the terminal knobs on the ends of the growing rRNA still attached to the ribosomal chromatin (r-chromatin) (Dragon et al. 2002).
Often two different steps in gene expression are coordinated. One example is the linkage between RNA polymerase II transcription and pre-mRNA processing. The physical association of pre-mRNA splicing and polyadenylation factors with the transcription apparatus enables binding of these RNA processing factors to the nascent mRNA transcript. It is not yet known whether there is a similar link between RNA polymerase I transcription and pre-rRNA processing. Indeed, it is not understood what triggers SSU processome association with the pre-rRNA, and what the order of component association is.
Here we present evidence that there is indeed coordination between pre-rRNA processing and RNA polymerase I transcription in yeast. We have found that some, but not all, of the SSU processome proteins are required for optimal pre-rRNA transcription in vivo and are closely associated with the r-chromatin. This subgroup of SSU processome proteins, the t-Utps, forms a complex on its own in the absence of the U3 snoRNA. In the absence of rDNA transcription, the subcomplex persists and is r-chromatin-associated. The t-Utps are associated with precursors to the 18S rRNA even when the U3 snoRNA is depleted, suggesting that their pre-rRNA association may represent an early step in the assembly of the SSU processome. Taken together, our results provide evidence that transcription of the rRNA precursor and its subsequent processing can be physically linked through this subset of SSU processome components.
Results
Transcription of the rDNA is reduced when a subset of Utps is depleted
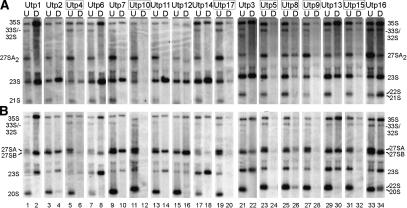
To carry out a comprehensive analysis of the function of the SSU processome components, we genetically depleted them to examine the effects of their absence on pre-rRNA processing. We used the GAL::HA-UTP strains that we had previously constructed and characterized (Dragon et al. 2002), in which each UTP is under the control of a galactose-inducible, glucose-repressible promoter. RNA was harvested from cells before and after depletion of Utp proteins, and the levels of the pre-rRNA precursors were determined by Northern blot analysis. Depletion of most of the Utps (Utp1, Utp2, Utp3, Utp6, Utp7, Utp11, Utp12, Utp13, and Utp14) led to deficient processing at A0, A1, and A2 (Fig. 2A, lanes 1–4,7–10,13–18,21,22,29,30). The 23S pre-rRNA, a normal intermediate resulting from cleavage at A3, accumulated, indicative of decreased cleavage at A0, A1, and A2. There was a concomitant loss of 27SA2 (indicating loss of cleavage at A2) and of the 21S pre-rRNA (A1 to A3; Lee and Baserga 1997; Torchet and Hermann-Le Denmat 2000). In addition, the 20S pre-rRNA, resulting from cleavage at A0, A1, and A2, and the precursor of 18S, was absent but the 27SB rRNA was present (Fig. 2B, lanes 1–4,7–10,13–18, 21,22,29,30). This pattern of precursors has been described many times for strains defective in proteins involved in SSU biogenesis (Jansen et al. 1993; Dunbar et al. 1997) and is characteristic of mutants defective in known U3-associated proteins. We will refer to these proteins as classical Utps, because their depletion leads to loss of A0, A1, and A2 cleavage events. As previously reported, levels of the U3 snoRNA were undiminished when classical Utps were depleted (Supplementary Fig. 1 in Dragon et al. 2002).
Figure 2.
Depletion of different components of the SSU processome leads to varying affects on steady-state pre-rRNA levels. Seventeen strains were constructed in which each Utp was expressed from a conditional (GAL) promoter (GAL::HA-UTP; Dragon et al. 2002). RNA was harvested from strains either prior to (undepleted [U]) or after (depleted [D]) depletion, as by Dragon et al. (2002). Following gel electrophoresis, the RNA was analyzed by Northern blots with specific oligonucleotides. The boxed names indicate the subgroup of Utps whose depletion affects levels of all pre-rRNAs (t-Utps). (A) Northern blots were probed with oligo c (between A2 and A3; Fig. 1). The expected precursors are labeled. (B) Northern blots were probed with oligo b (between A1 and A2; Fig. 1) and oligo e (in ITS2; Fig. 1).
Utp16 (Bud21) is so far the only nonessential protein in the SSU processome; however, depletion of Utp16 causes cold sensitivity (Dragon et al. 2002). Examination of the resulting precursors from cells grown at room temperature on Northern blots indicated a defect in the processing at A1 and A2 because the levels of 27SA2 were reduced (Fig. 2A, lanes 33,34) and because the 22S (A0–A3) pre-rRNA accumulated (Fig. 2B, lanes 33,34). Accumulation of this precursor has been observed in mutants in the U3 snoRNA and in an mpp10 mutant that confers cold sensitivity, and following depletion of Dhr1 (Hughes 1996; Lee and Baserga 1997; Colley et al. 2000).
Unexpectedly, depletion of a subset of Utps (Utp4, Utp5, Utp8, Utp9, Utp10, Utp15, and Utp17) led to a gross reduction of all the rRNA precursors to both large and small ribosomal subunits when examined by steadystate analysis. Although the extent was variable, reductions in levels of the primary transcript, the 35S, and the 33S/32S, 27SA, 27SB, 23S, 21S, and 20S pre-RNAs were observed (Fig. 2A,B, lanes 5,6,11,12,19,20,23–28,31,32). This was in contrast to the findings obtained following depletion of the classical Utps described above. However, we could not attribute this to general RNA degradation or experimental loss of RNA, because the levels of the U3 snoRNA remained undiminished, as previously reported (Supplementary Fig. 1 in Dragon et al. 2002). This result was surprising, as these Utps are clearly SSU processome components: They associate with the U3 snoRNA and Mpp10, localize to the nucleolus, and are required for 18S rRNA maturation (Dragon et al. 2002), and we expected their depletion to result in the previously described pre-rRNA processing defects for U3-snoRNA associated proteins, like those observed in Figure 2 for the classical Utps.
One explanation for the loss of all pre-rRNAs upon depletion of this subset of Utps is that they are required for optimal transcription of the rDNA by RNA polymerase I. This would result in a reduction in the levels of the pre-rRNAs by steady-state RNA analysis. We tested this in two ways: by transcription run-on analysis and by quantitation of the number of rRNA transcripts per gene in chromatin spreads in electron microscopy. To avoid secondary defects on transcription due to the prolonged absence of ribosomes, the experiments were carried out soon after the beginning of protein depletion.
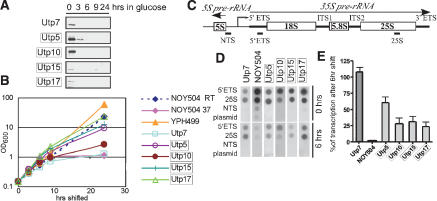
Strains expressing a tagged classical Utp (Utp7) or nonclassical Utps (Utp5, Utp10, Utp15, and Utp17) were expressed from a galactose-inducible, glucose-repressible promoter in strains (GAL::HA-UTP) where they are each the sole source of these essential proteins. At 6 h after the switch to glucose, protein levels for Utp7, Utp5, Utp10, Utp15, and Utp17 were substantially reduced, indicating their depletion (Fig. 3A). However, during the same time period there was not yet an appreciable change in growth (Fig. 3B), indicating that ribosomes had not yet become limiting for cell growth. We compared the results with these strains to those obtained with strain NOY504, which bears a temperature-sensitive RNA polymerase I that effectively results in depletion of a major essential subunit at the nonpermissive temperature (Nogi et al. 1991). It was previously shown that after 6 h at the nonpermissive temperature, transcription ceases in this strain, whereas growth is not yet affected (Fig. 3B).
Figure 3.
Transcription run-on analysis indicates that depletion of a subgroup of SSU processome components leads to a reduction in pre-rRNA transcription. Each strain carries the single gene encoding the indicated protein under the control of a GAL promoter (GAL::HA-UTP), allowing for conditional expression. The boxes indicate proteins whose depletion affects levels of all pre-rRNAs (from Fig. 2), and that were therefore hypothesized to be affecting transcription. The NOY504 strain carries a temperature-sensitive RNA polymerase I. (A) The Utp proteins are depleted when the GAL::HA-UTP strains are grown in glucose. Western blots with anti-HA antibody were performed on protein harvested from yeast expressing the indicated tagged proteins under the control of a GAL promoter prior to (0) or after (3, 6, 9, 24 h) the switch to glucose. (B) Growth curves of yeast strains. Growth of the GAL::HA-UTP and the parent YPH499 strains in glucose is indicated graphically and compared to growth of the NOY504 strain at the permissive (room temperature [RT]) and nonpermissive (37°C) temperatures. (C) The rDNA transcription unit. RNA polymerase I transcribes the 35S primary rRNA transcript, whereas RNA polymerase III transcribes the 5S rRNA in the opposite direction. PCR products corresponding to the indicated segments of the nontranscribed spacer (NTS), the 5′ external transcribed spacer (5′ETS), and the 25S rRNA were cloned into the pTOPO plasmid. (D) Transcription run-on analysis. The GAL::HA-UTP strains were either not depleted or depleted for 6 h of the indicated proteins, permeabilized, and exposed to α32P-UTP for 10 min. RNA was harvested and hybridized to plasmids described in C dot-blotted onto Hybond-N+ membrane. For the NOY504 strain, cultures were incubated at 37°C for 6 h. (E) Quantitation of the results in D.
Transcription run-on analysis has been used to quantify the transcriptional activity of genes, including the rDNA, in vivo (Elion and Warner 1986; Weston and Bishop 1989) because it provides a snapshot of the transcripts that have initiated transcription when the label is added. Unlike with other methods, transcription reinitiation does not occur. We performed transcription run-on analysis of yeast on cells depleted for 6 h of the classical Utp, Utp7, and the nonclassical Utps, Utp5, Utp10, Utp15 and Utp17. We compared the resulting changes in transcription to the changes observed in the NOY504 strain over the same time period. Yeast were permeabilized and incubated with α-32P-UTP for 10 min. The labeled RNAs were isolated and hybridized to a dot blot containing plasmids with DNA sequences (Fig. 3C) representing transcribed portions of the rDNA (5′ETS, 25S), the nontranscribed spacer (NTS), and to plasmid DNA as a control for nonspecific hybridization. For the transcription run-ons to be quantitative, the amount of DNA on the dot blot must be in excess of the maximum levels of RNA transcripts. This was verified prior to proceeding with the depletion studies (data not shown). As was previously observed, 6 h at the nonpermissive temperature greatly reduced transcription in the NOY504 strain (Fig. 3D; Nogi et al. 1991). Similarly, depletion of the nonclassical Utps, Utp5, Utp10, Utp15, and Utp17 for 6 h also resulted in decreased transcription (Fig. 3D). In contrast, no changes in transcription upon depletion of the classical Utp, Utp7, were observed (Fig. 3D). The results from two experiments were quantitated and are shown in Figure 3E. From this bar graph it is clear that transcription decreases greatly when RNA polymerase I is depleted but does not change when Utp7 is depleted. Depletion of each of Utp5, Utp10, Utp15, and Utp17 resulted in reduced transcription, though not to as great an extent as that observed upon depletion of RNA polymerase I. Our results therefore indicate that depletion of the nonclassical Utps leads to a decrease in the number of rRNA transcripts, strongly suggesting that they are required for optimal transcription. Because the transcription run-on assays were carried out on cells that were still growing normally, it is likely that the effect that we observe on transcription is a direct result of depletion of these particular proteins. We will thus refer to this subset of SSU processome proteins as t-Utps, for U3 proteins (Utp) required for transcription (t).
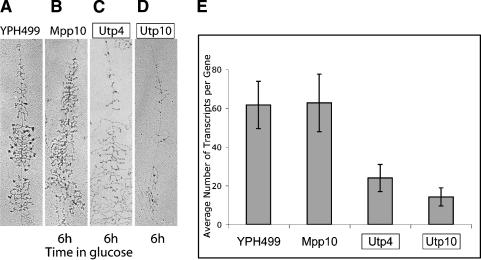
Transcription of the rDNA can also be directly assessed by counting transcripts on the rRNA genes in chromatin spreads (Miller spreads) with an electron microscope. This number is equal to the number of RNA polymerases per gene in the process of transcribing the rDNA. The number of transcripts per gene was obtained following depletion of Mpp10, a classical Utp, and two t-Utps, Utp4 and Utp10, and compared to the number obtained for the parent strain, YPH499. The GAL::MPP10, GAL::UTP4, GAL::UTP10, and YPH499 strains were grown in glucose for 6 h. The YPH499 strain yielded the expected highly transcribed genes with characteristic large terminal knobs (Fig. 4A). Depletion of Mpp10 yielded no significant change in transcript density, though terminal knobs were lost as shown previously for other SSU processome components (Fig. 4B; Dragon et al. 2002). In contrast, depletion of both Utp4 and Utp10 yielded transcription units with very sparse transcripts (Fig. 4C,D). The number of rRNA transcripts per gene was quantitated by counting transcripts on 32 genes for each strain and averaging them. The results are depicted in the histogram in Figure 4E. The parent strain, YPH499, had 61.6 transcripts/gene (standard deviation [STD] 12.2). Depletion of Mpp10, a classical Utp, resulted in a similar number of transcripts/gene, 62.7 (STD 14.8). Thus, depletion of Mpp10, as expected, does not affect transcription of the rDNA. In contrast, depletion of the t-Utps resulted in many fewer transcripts per gene. Depletion of Utp4 resulted in 23.9 transcripts per gene (STD 7.0) whereas depletion of Utp10 resulted in 14.2 transcripts per gene (STD 4.7). The t-test indicates that the difference between the t-Utp strains and the other strains is highly significant (P < 0.001). Therefore, analysis of chromatin spreads by electron microscopy corroborates our results obtained by transcription run-on: that the t-Utps are required for optimal transcription of the rDNA.
Figure 4.
Quantitation of the number of rRNA transcripts per gene in chromatin spreads indicates that depletion of the t-Utps leads to a reduction in pre-rRNA transcription. Each strain carries the gene encoding the indicated protein under the control of a GAL promoter, allowing for conditional expression. The parent strain is YPH499. The boxes indicate t-Utps. (A–D) Examples of chromatin spreads from the indicated strains. The Mpp10, Utp4, and Utp10 proteins were depleted by growth in glucose for 6 h. (E) Results from quantitation of the number of rRNA transcripts per gene upon depletion of the indicated proteins. The average number of transcripts per gene for 32 genes for each strain is shown ± standard deviation.
The t-Utps are associated with the r-chromatin
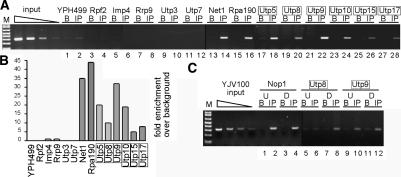
In light of the role of a subset of Utps in transcription, we examined the ability of HA-tagged t-Utps and other nucleolar proteins to interact with the r-chromatin (rDNA with associated proteins) using chromatin immunoprecipitation (Gotta et al. 1997; Rusche and Rine 2001; Strasser et al. 2002). Following in vivo cross-linking, shearing of the DNA, and immunoprecipitation with tagged proteins, chromatin association was revealed by using polymerase chain reaction (PCR) to detect a fragment of the 5′ETS. The results indicated that, as expected, a subunit of RNA polymerase I, Rpa190, and an associated protein, Net1 (Shou et al. 2001), interacted with the r-chromatin (Fig. 5A, lanes 14,16), whereas there was no signal with the untagged parental strain, YPH499 (Fig. 5A, lane 2). The SSU processome proteins fell into two exclusive categories: The t-Utps (Utp5, Utp8, Utp9, Utp10, Utp15, and Utp17) did associate with the r-chromatin (Fig. 5A, lanes 18,20,22,24,26,28), whereas the classical Utps (Imp4, Rrp9, Utp3, and Utp7) did not (Fig. 5A, lanes 6,8,10,12). Two members of the Imp4 superfamily of RNA-binding proteins, Imp4 and Rpf2, which have in vitro RNA-binding capabilities (Wehner and Baserga 2002), were also found not to associate with the r-chromatin, demonstrating that association with the r-chromatin was not a general result of the nucleic acid affinity of these proteins (Fig. 5A, lanes 4,6). Quantitation with normalization to the amount of immunoprecipitate bound to beads without antibody (beads-alone lanes, marked B) indicates that Rpa190, Net1, and the t-Utps are all associated with the r-chromatin (Fig. 5B). In contrast, the classical Utps and two Imp4 superfamily proteins are not r-chromatin-associated (Fig. 5B). Therefore, the t-Utps are required for optimal rDNA transcription and are closely associated with the r-chromatin.
Figure 5.
The t-Utps are closely r-chromatin-associated. The indicated proteins were HA-tagged by integration of the tag at the genomic locus. YPH499 is the parental, untagged strain. Chromatin immunoprecipitations (ChIPs) with anti-HA antibodies (IP) (except for Rpa190, which was immunoprecipitated via the TAP tag with IgG beads) or with beads alone (B) were performed as described in Materials and Methods. R-chromatin association was assessed by assaying for the presence of immunoprecipitated 5′ETS sequences using PCR. The t-Utps are boxed. (A) ChIPs of tagged proteins required for ribosome biogenesis. Rpf2 and Imp4 are RNA-binding proteins containing the σ70-like motif, and are required for LSU and SSU biogenesis, respectively. Imp4, Rrp9, Utp3, and Utp7 are components of the SSU processome whose depletion does not affect transcription. Net1 is an RNA polymerase I-associated protein, and Rpa190 is a subunit of RNA polymerase I; they were used here as positive controls. Utp5, Utp8, Utp9, Utp10, Utp15, and Utp17 are t-Utps. (B) Quantitation of the results in A. (C) The t-Utps persist in their r-chromatin association in the absence of rRNA transcription. The indicated proteins were HA-tagged in the YJV100 strain by chromosomal integration. In the YJV100 strain, the endogenous rDNA has been replaced with 20–25 copies of the rDNA under the control of a galactose-inducible, glucose-repressible (GAL) Pol II promoter (Venema et al. 1995). rRNA transcription was interrupted (7 h, D) or not (0 h, U) by growth in glucose. ChIP was performed with immunoprecipitations with anti-HA antibodies (IP) or beads alone (B). Coimmunoprecipitating rDNA was analyzed by PCR with primers specific to portions of the 5′ETS.
Is transcription required for r-chromatin association of the t-Utps?
During transcription of pre-mRNA, splicing and polyadenylation factors are brought to the nascent mRNA by RNA polymerase II (for review, see Howe 2002; Proudfoot and O'Sullivan 2002). By analogy, if RNA polymerase I were bringing the t-Utps, transcription would be required for their r-chromatin association. To test this, transcription was down-regulated in a strain where rDNA transcription could be manipulated. In yeast, the rDNA can be expressed from an RNA polymerase II promoter (Nogi et al. 1991). In the YJV100 strain, the chromosomal rDNA with its RNA polymerase I promoter has been deleted and replaced with 20–25 copies of the rDNA under the control of a galactose-inducible, glucose-repressible (GAL) Pol II promoter (Venema et al. 1995). The rDNA is therefore transcribed in galactose but not in glucose. Two randomly selected t-Utps (Utp8, Utp9) and Nop1 were epitope-tagged with HA by chromosomal integration in YJV100. RNA polymerase II transcription of the rDNA was interrupted by growth in glucose for 7 h. For this strain, 7 h in glucose is sufficient to cause loss of rDNA transcription, but cell growth is not yet affected due to the excess of ribosomes present (Venema et al. 1995). Association with the r-chromatin was assessed before and after transcription was turned off. R-chromatin association of the t-Utps occurred even when the rDNA was transcribed from an RNA polymerase II promoter (Fig. 5C, 0 h, lanes 2,6,10), and it persisted after transcription ceased (Fig. 5C, 7 h, lanes 4,8,12). These results suggest that the presence of the rDNA is sufficient for association of the t-Utps with the r-chromatin. In addition, the results rule out the possibility that the observed r-chromatin association occurs by binding of the t-Utps to the nascent rRNA. Furthermore, they indicate that transcription by RNA polymerase I is not required for r-chromatin association of the t-Utps, and, more importantly, that even transcription itself is not required.
Are the t-Utps found in a complex in the absence of the SSU processome?
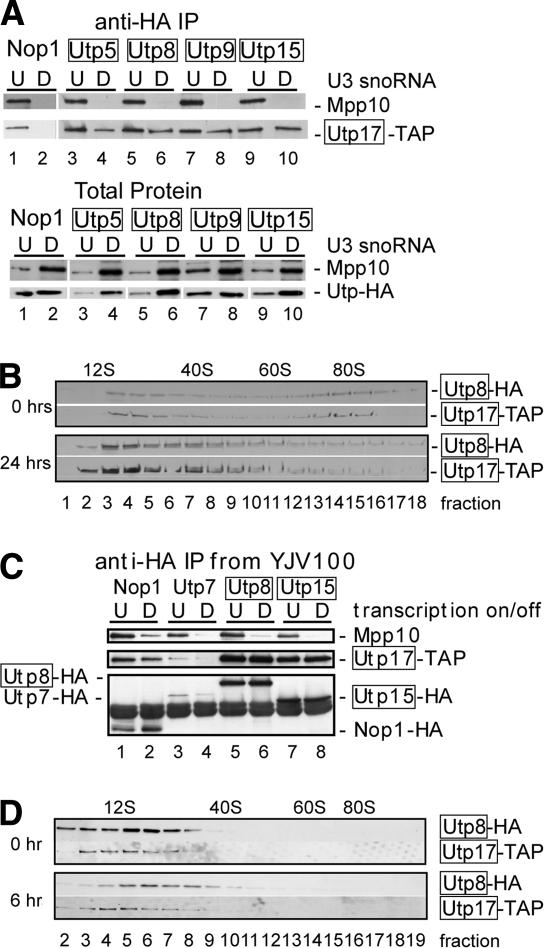
Because they have a similar function in transcription and because they were present on the r-chromatin, we hypothesized that the t-Utps might be present in a complex in the absence of the SSU processome, perhaps representing an earlier complex in SSU biogenesis. To assess this, we reduced levels of the SSU processome by depletion of the U3 snoRNA. This had previously been shown to result in loss of the SSU processome as visualized by the absence of terminal knobs on the ends of nascent rRNAs (Dragon et al. 2002). Proteins were either HA- or TAP-tagged, as indicated, in a strain where the U3 snoRNA could be depleted by growth in glucose (YKW100; Wehner et al. 2002). The YKW100 strain was derived from JH84, and bears a disruption of one of the U3 snoRNA genes, whereas the other is under control of the galactose (GAL) promoter (Hughes and Ares 1991; Samarsky and Fournier 1998). This strain permits depletion of the U3 snoRNA by growth in glucose. As expected, in cells not depleted of the U3 snoRNA (growth in galactose; 0 h), the SSU processome components Nop1, Utp5, Utp8, Utp9, and Utp15 coimmunoprecipitated Mpp10 and Utp17, indicating the presence of the SSU processome (Fig. 6A, lanes 1,3,5,7,9). However, in cells depleted of the U3 snoRNA (growth in glucose; 24 h), the SSU processome components Nop1, Utp5, Utp8, Utp9, and Utp15 no longer coimmunoprecipitated Mpp10, indicating disruption of the SSU processome (Fig. 6A, top, lanes 2,4,6,8,10). Similarly, Nop1 did not coimmunoprecipitate Utp17 when the U3 snoRNA was depleted (Fig. 6A, top, lane 2). The lack of coimmunoprecipitation is not due to a reduction in protein levels upon U3 snoRNA depletion, because levels of the SSU processome proteins remained largely undiminished (Fig. 6A, bottom). In contrast, randomly selected t-Utps, Utp5, Utp8, Utp9, and Utp15 all persisted in coimmunoprecipitating another t-Utp, Utp17, when the U3 snoRNA was depleted (Fig. 6A, top, lanes 4,6,8,10). This indicates that the SSU processome is not required for formation of a complex that includes the t-Utps.
Figure 6.
(A) The t-Utps remain in a complex in the absence of the SSU processome. Proteins in the YKW100 strain were tagged with both the HA (Nop1, Utp5, Utp8, Utp9, and Utp15) and TAP (Utp17) tags, as indicated, by integration at the genomic locus. The t-Utps are boxed. The U3 snoRNA was either not depleted (0 h) or depleted (24 h). Anti-HA immunoprecipitation was performed on extracts from either depleted (D) or undepleted (U) cells, and proteins were analyzed by Western blot with anti-Mpp10 antibodies (to detect Mpp10) and with PAP (to detect the TAP tag on Utp17; top panel). (Bottom panel) Depletion of the SSU processome does not affect levels of SSU processome proteins. The U3 snoRNA was depleted (24 h, D) or not (0 h, U), and total protein levels were analyzed by Western blot with anti-Mpp10 antibodies and anti-HA antibodies (to detect the HA-tagged proteins). (B) Sedimentation profile of the t-Utps upon depletion of the SSU processome. Extracts from an HA-Utp8, TAP-Utp17-tagged YKW100 strain where the U3 snoRNA was either depleted (24 h) or not (0 h) were run on 10%–47% sucrose gradient. Fractions were analyzed by Western blot with anti-HA antibodies (to detect Utp8) or PAP (to detect Utp17). (C) The t-Utps remain in a complex in the absence of rRNA transcription. The indicated proteins were HA-tagged in the YJV100 strain by chromosomal integration. In the YJV100 strain, the endogenous rDNA was replaced with 20–25 copies of the rDNA under the control of a galactose-inducible, glucose-repressible (GAL) Pol II promoter. rRNA transcription was interrupted (7 h, D) or not (0 h, U) by growth in glucose. Anti-HA immunoprecipitations were performed on cell extracts, and coimmunoprecipitating proteins were analyzed by Western blot with anti-Mpp10 and anti-HA antibodies, and with PAP (to detect the TAP tag on Utp17). (D) Sedimentation profile of the t-Utps before and after interruption of transcription. Extracts from an HA-Utp8, TAP-Utp17-tagged YJV100 strain where shift to glucose (6 h) interrupted transcription were run on 10%–50% sucrose gradient. Fractions were analyzed by Western blot with anti-HA antibodies (to detect Utp8) or PAP (to detect Utp17).
Total cellular extracts from cells depleted of the SSU processome were analyzed on 10%–47% sucrose gradients to assess the size of the t-Utp subcomplex. In this strain, two t-Utps, Utp8 and Utp17, were HA- and TAP-tagged, respectively, in a strain where the U3 snoRNA can be depleted (YKW100). As indicated by Western blotting of the gradient fractions, the complex sedimented in particle sizes of ∼20S and 80S (SSU processome) in undepleted cells (Fig. 6B, 0 h). Following depletion of the SSU processome, the t-Utps were largely in the 20S-sized complex (Fig. 6B, 24 h). It is likely that this represents a soluble t-Utp complex, and not the complex associated with the r-chromatin. Thus, when the SSU processome is depleted, the t-Utp subcomplex remains intact.
We also examined whether the t-Utps remain in a complex in the absence of transcription. We tagged examples of the relevant proteins in the YJV100 strain, where we can regulate rDNA transcription by carbon source. This strain carries a deletion of the usual rDNA transcription unit that has been replaced with an rDNA locus consisting of 20–25 copies of the transcription unit under the control of a galactose-inducible (GAL) promoter. A YJV100-derived strain was constructed in which Utp17 (t-Utp) was TAP-tagged. Nop1, Utp7 (classical Utps), and Utp8 and Utp15 (t-Utps) were then HA-tagged in this strain. Coimmunoprecipitations were performed between Nop1, Utp7, the t-Utps, and Mpp10 (classical Utp) or a TAP-tagged Utp17. Noticeably, lack of rDNA transcription led to loss of coimmunoprecipitation of SSU processome components, Nop1, Utp7, Utp8, and Utp15 with Mpp10 (Fig. 6C, top panel, lanes 2,4,6,8). This indicates that in the absence of transcription the SSU processome does not form. In contrast, in the absence of transcription, coimmunoprecipitation between t-Utps persisted (Utp8 or Utp15 with Utp17), indicating that, in the absence of transcription, this subcomplex is extant (Fig. 6C, middle panel, lanes 6,8). In all cases the HA-tagged protein was present in each immunoprecipitation, and therefore the lack of coimmunoprecipitation was not due to protein degradation (Fig. 6C, bottom panel). The t-Utps are thus associated with each other both in the absence of transcription and in the absence of the SSU processome.
To assess the size of the t-Utp subcomplex before and after interruption of transcription, total cellular extracts were analyzed on 10%–50% sucrose gradients. In this YJV100 strain, two t-Utps, Utp8 and Utp17, are HA- and TAP-tagged, respectively. As indicated by Western blotting of the gradient fractions, the soluble complex sedimented in a particle size of ∼20S in yeast before and after transcription had ceased (Fig. 6D, 0 and 6 h), indicating no change in the size of the t-Utp subcomplex upon depletion. Thus, the t-Utp subcomplex remains intact in the absence of transcription. We do not know why we cannot detect the 80S SSU processome in this strain, but it is likely due to the reduced number of active transcription units that results in extreme slow growth of the yeast even prior to interruption of transcription.
The t-Utps are associated with pre-rRNAs
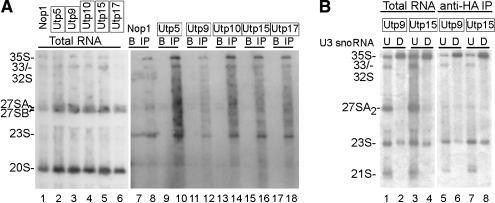
Because the t-Utps are components of the SSU processome, we asked with which rRNA precursors the proteins were associated. Randomly selected HA-tagged t-Utps, Utp5, Utp9, Utp10, Utp15, and Utp17 all coimmunoprecipitated 35S, 33S/32S, and the 23S pre-rRNA (Fig. 7A, lanes 10,12,14,16,18). This is identical to our previous results with the SSU processome component Imp4 (Wehner and Baserga 2002). Likewise, Nop1p coimmunoprecipitated the 35S, 33S/32S, 27SA, 27SB, and 23S rRNAs, consistent with its role in methylation of all pre-rRNA species and in the SSU processome (Fig. 7A, lane 8). Thus, the t-Utps are associated with the same pre-rRNAs as the classical Utps. These results also demonstrate that although t-Utp depletion also affects the levels of the precursors containing the LSU rRNAs, they do not directly interact with them and therefore are likely not to have a direct role in their maturation after transcription. This suggests that the reduction in levels of the LSU rRNAs visualized on Northern blots results from reduced transcription alone.
Figure 7.
(A) The t-Utps coimmunoprecipitate pre-18S rRNAs. The indicated proteins were HA-tagged by integration of the tag at the genomic locus. Boxes indicate t-Utps. “B” indicates immunoprecipitations performed with beads alone (no antibody), “IP” indicates immunoprecipitations performed with anti-HA antibody. Coimmunoprecipitating RNAs were isolated and analyzed by Northern blot with oligonucleotides complementary to the pre-rRNAs (oligos c, b, and e; Fig. 1). Pre-rRNA species were determined by their size relative to total RNA extracted by the hot phenol method from the parental YPH499 strain (data not shown). (B) The t-Utps coimmunoprecipitate pre-18S rRNAs in the absence of the U3 snoRNA. The indicated t-Utps were HA-tagged in the YKW100 strain. Anti-HA immunoprecipitations were performed on extracts from yeast depleted (D) or not (U) of the U3 snoRNA by growth in glucose. Coimmunoprecipitating RNAs were isolated and analyzed by Northern blot with an oligonucleotide complementary to pre-rRNAs (oligo c; Fig. 1).
To order the association of the t-Utps and the U3 snoRNA on the pre-rRNAs, we asked whether the t-Utps were associated with pre-rRNAs in the absence of the SSU processome. Immunoprecipitations of HA-tagged t-Utps were performed on yeast depleted of the U3 snoRNA. The associated pre-rRNAs were examined by Northern blotting. Immunoprecipitation of HA-tagged tUtps, Utp9, and Utp15 indicated that their association with the primary 35S transcript and the 23S rRNA intermediate persisted when the U3 snoRNA was depleted (Fig. 7B, lanes 6,8). Thus, the t-Utps do not require formation of the SSU processome for their pre-rRNA association. This supports an order of assembly for the SSU processome where the t-Utps assemble on the pre-rRNA prior to the U3 snoRNA.
Discussion
The SSU processome is a large RNP, visualized as the terminal knobs on nascent RNAs, that is required for processing of pre-18S rRNAs. A subset of SSU processome components, the t-Utps, affected steady-state levels of all pre-rRNAs when they were depleted, compelling us to ask whether they were required for rDNA transcription. Indeed, transcription run-on analysis and quantitation of the number of rRNA transcripts/gene in chromatin spreads indicated that transcription was reduced when the t-Utps, but not the classical Utps, were depleted, suggesting strongly that the t-Utps are required for optimal transcription. Chromatin immunoprecipitation indicated that the t-Utps, but not the classical Utps, were associated with the r-chromatin, and that their association did not require transcription. Depletion of the U3 snoRNA indicated that the t-Utps were present in a subcomplex in the absence of the SSU processome. This subcomplex remained intact also in the absence of transcription. As expected for components of the SSU processome, the t-Utps were found associated with precursor RNAs to the 18S rRNA but not with the direct precursors to the LSU rRNAs. The t-Utps were also associated with the 35S and 23S pre-rRNAs in the absence of the U3 snoRNA, indicating that the presence of the SSU processome is not required for their association with these transcripts. Thus, these results provide strong evidence for a link between rDNA transcription and pre-rRNA processing by components of the SSU processome.
The identification of a subset of SSU processome components, the t-Utps, as having a role in pre-rRNA transcription was unexpected. The t-Utps were originally identified because they copurified with the SSU processome (Dragon et al. 2002). Like other SSU processome components, they are nucleolar, and they coimmunoprecipitate the U3 snoRNA and another SSU processome component, Mpp10. Pre-rRNA immunoprecipitation with the t-Utps indicates that they are present in the SSU processome even after the SSU pre-rRNA is separated from the LSU pre-rRNA by cleavage at A3. By these criteria they are similar to the other tested components of the SSU processome.
However, the t-Utps differ from the other SSU processome components in a number of ways. We first detected them in analyzing the pre-rRNA processing defects resulting from SSU processome protein depletion. The depletion of most SSU processome components, including the U3 snoRNA, results in a predictable pattern of pre-rRNA precursors on Northern blots caused by defective cleavage at the A0, A1, and A2 sites. In contrast, when analyzed in the same way, depletion of the t-Utps results in no particular RNA pattern but in even loss of all the pre-rRNAs. This result is supported by re-examination of our previously published results where, although 18S rRNA levels appeared to be more greatly affected, 25S rRNA levels were also decreased upon depletion of most of the t-Utps (Dragon et al. 2002). Transcription run-on analysis and direct quantitation of the number of rRNA transcripts per gene indicated that this was due to a reduction in pre-rRNA transcription that was not observed upon depletion of other SSU processome components. In addition, the t-Utps but not the other tested SSU processome components are associated with the r-chromatin. The t-Utps also remain associated with each other in the absence of the SSU processome. Therefore, although the t-Utps are components of the SSU processome, they differ from other SSU processome components in several ways and thus contribute in a unique way to ribosome biogenesis.
Prior to this study, no protein component of any large pre-ribosomal RNP had been shown to be required for optimal transcription. However, studies in Xenopus oocytes demonstrated that excess injected nucleolin, an abundant nucleolar protein with several proposed functions, interfered with both pre-rRNA transcription and maturation (Roger et al. 2002, 2003). It is not clear how excess nucleolin interferes with these two aspects of ribosome synthesis.
Our results suggest that the presence of the rDNA itself is sufficient for the r-chromatin association of the t-Utps because neither the transcribed rRNA nor the presence of RNA polymerase I is required. Supporting our conclusion that the r-chromatin association of the t-Utps is independent of transcription, we have not been able to demonstrate significant increased coimmunoprecipitation of RNA polymerase I subunits and this subset of SSU processome components compared to coimmunoprecipitation of RNA polymerase I subunits and other SSU processome components (data not shown). This is distinct from the way that RNA polymerase II recruits both pre-mRNA splicing and polyadenylation factors by direct contact (for review, see Howe 2002; Proudfoot and O'Sullivan 2002). Therefore, although transcription and RNA processing are linked for production of rRNAs, as transcription and RNA processing are linked for production of mRNAs, it is through a different mechanism.
Recently Krogan et al. (2004) undertook an analysis of RNA-related processing complexes in yeast and also identified the t-Utp complex, which they termed UTPA. Their aim was to isolate complexes smaller than the ribosome (80S) by performing an 180,000g centrifugation on extracts prior to purification with the tagged protein. UTPA has seven members: Utp4, Utp8, Utp9, Utp10, Utp15, Utp17/Nan1, and Pol5. Our t-Utp complex, identified as components of the SSU processome, also has seven members: Utp4, Utp5, Utp8, Utp9, Utp10, Utp15, and Utp17/Nan1. Thus the complexes are almost the same with the exception that their complex contains the protein Pol5 whereas ours contains Utp5. Pol5 is a protein so named because it bears sequence similarity to DNA polymerases and has DNA polymerase activity in vitro (Shimizu et al. 2002). However, its DNA polymerase activity is not required for its essential function in vivo. Furthermore, it resides in the nucleolus and is required for ribosome biogenesis. We have performed transcription run-on assays in a strain depleted of Pol5 and found that it is required for rDNA transcription (data not shown). Chromatin immunoprecipitation indicates that Pol5 is associated with the r-chromatin (data not shown). However, association of Pol5 with other SSU processome components was low by coimmunoprecipitation (data not shown), suggesting that it may be present in substoichiometric amounts in the SSU processome or present in only very early complexes. Although Krogan et al. (2004) did not assign a function in transcription to the UTPA complex, their analyses, carried out very differently from ours, yielded some similar findings. Inspection of their results upon UTPA protein depletion indicated a decrease in levels of both SSU and LSU pre-rRNAs, consistent with the results presented here in Figure 2.
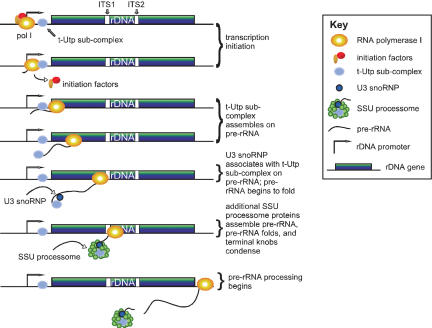
The results presented here are consistent with a model for SSU processome assembly that begins in an unanticipated way with the t-Utps associated with the rDNA itself (Fig. 8). In this model we have indicated t-Utp association with the 5′ETS, but we cannot rule out that association also occurs with other portions of the transcription unit. As the pre-rRNA is transcribed by RNA polymerase I, the t-Utps associate with the nascent transcript. The U3 snoRNP associates with the subcomplex on the nascent rRNA transcript, U3 snoRNA:pre-rRNA base-pairing occurs, and the pre-rRNA begins to fold. As transcription through the 18S rRNA is completed, the additional SSU processome proteins associate with the subcomplex/U3 snoRNP to form the SSU processome. This large RNP can be visualized by electron microscopy as the terminal knobs on nascent rRNAs in chromatin spreads. Once the SSU processome is assembled, pre-18S rRNA processing can then proceed.
Figure 8.
Model linking rRNA transcription and pre-rRNA processing via components of the SSU processome. Association of the t-Utps with the r-chromatin is required for efficient transcription, and allows their assembly onto the nascent rRNA transcript. The U3 snoRNP then associates with the rRNA via base-pairing interactions, additional proteins join, and the SSU processome is formed. As a result, the pre-18S rRNA becomes accurately folded and ready for rRNA processing.
This model predicts that some or all of the t-Utps are both rDNA- and rRNA-binding proteins. This dual nucleic acid binding capacity is not without precedent as there are a number of proteins, many of which are transcription factors, which bind both DNA and RNA (for review, see Cassiday and Maher 2002). The prototypical example is TFIIIA, discovered both as an activator of 5S rRNA transcription, where it binds to DNA, and as the storage protein for the 5S rRNA in Xenopus oocytes. Further experimentation will determine whether some or all of the t-Utps indeed belong to the growing list of proteins with an interesting dual function in DNA and RNA binding.
Materials and methods
Strains and media
Strains containing triple HA carboxyl-tagged proteins (KanR), galactose-inducible triple HA amino-tagged proteins (GAL::HA-UTP or otherwise; KanR) and TAP carboxyl-tagged proteins (Kluyveromyces lactis TRP1 in YPH499 and YKW100 or S. cerevisiae HIS3 in YJV100) were constructed as described by Dragon et al. (2002). YPH499 was used as a control strain and the parental strain unless otherwise noted. YKW100 was described by Wehner et al. (2002), NOY504 by Nogi et al. (1991), and YJV100 by Venema et al. (1995). All strains were grown as outlined by Wehner et al. (2002). YPH499 and NOY504 were grown in YPD (1% yeast extract, 2% peptone, 2% dextrose). YJV100, YKW100, and GAL::HA-UTP strains were grown in YPG/R (1% yeast extract, 2% peptone, 2% galactose, and 2% raffinose) until shifted to YPD for the indicated time period to down-regulate expression of the genes under the control of the GAL promoter. NOY504 was grown at 22°C and then shifted to 37°C for the indicated time. All other yeast strains were grown at 30°C.
RNA manipulations
Depletion of the Utps for steady-state RNA analysis (Fig. 2) from GAL::HA-UTP strains was carried out as described by Dragon et al. (2002), for either 24 or 48 h, depending on when growth changed. Harvesting of RNA and Northern blots were carried out according to Dunbar et al. (1997). To analyze highmolecular-weight RNAs, equal amounts of total RNA were loaded in each lane and separated on formaldehyde-1.25% agarose gels. Oligonucleotides b, c, and e used for Northern blotting were characterized by Beltrame and Tollervey (1992). The method for pre-rRNA coimmunoprecipitation with HA-tagged proteins (Fig. 7) was described by Wehner and Baserga (2002). Extract for RNA coimmunoprecipitations was made from an equal number of cells.
Transcription run-on assays
GAL::HA-UTP strains were grown in liquid YPG/R. Three mL of cells at an O.D.600 of 0.4–0.5 was harvested after a 6-h shift to YPD. Transcription run-on assays were performed according to the protocol of Elion and Warner (1986) with the following changes: The cells were incubated with α-32P-UTP for 10 min and the RNA was extracted according to the method of Ausubel et al. (1995). PCR fragments corresponding to segments of the rDNA transcription unit were cloned into the pTOPO plasmid, and 10 µg was spotted onto Hybond N+ membrane (Amersham Biosciences). The RNA was denatured and hybridized overnight to a dot blot according to the Transcription Run-on Protocol by J. Michael Bishop at Bio.com (http://www.bio.com). Primers to the 5′ETS (see ChIP methods for the sequences of the primers), 25S of the rDNA, and NTS were used to generate the PCR products for the dot blots. The sequences of the primers for the 25S rDNA are 5′ 25S-5270 (GACTACTTGCGTGCCTTGTTG) and 3′ oligo y (CCGTTCCCTTGGCTGTG). The sequences for the NTS primers are 5′ 5S (GGTAGATATGGCCGCAACC) and 3′NTS2 (CTCACACTTGTACTCCCATGAC). The parent plasmid, pTOPO, was used to assess nonspecific binding of labeled RNA. The dot blot hybridization signals were quantitated using Bio-Rad Multi-Analyst Version 1.0.2. To obtain the corrected values, the background (the value of the NTS dot) was subtracted from the 5′ETS and 25S dot values. The percentage decrease of transcription after 6 h of shift was calculated by dividing the corrected value at 0 h by the corrected value at 6 h. The percent decrease from two independent transcription run-on assays was averaged together and standard error is indicated.
Chromatin (Miller) spreads
Yeast cultures were grown in YP medium plus galactose, raffinose, and 1 M sorbitol to an absorbance at 600 nm of 0.4. For depletions, the cultures were split, spun down, washed in appropriate medium, respun, and resuspended in either YP medium plus galactose, raffinose, and 1 M sorbitol (controls) or YP medium plus glucose and 1 M sorbitol (depletions). Miller spreads of both preparations were made at 6 h postdepletion according to the method of Dragon et al. (2002). Briefly, 1 mL of culture was digested for 4 min at 30°C with 5 mg zymolyase which had been prewarmed in 200 µL of appropriate medium. Cells were spun down for 10 sec, and the pellet was resuspended in 1 mL of 0.025% Triton X-100 at pH 9. This solution was then diluted into 3 mL of the Triton solution and dispersed for 20 min with swirling. After addition of one-tenth volume of 0.1 M sucrose-10% formalin at pH 8.7, grids were made as described (Osheim and Beyer 1989).
Chromatin immunoprecipitations
The protocol was modified from Gotta et al. (1997) and Rusche and Rine (2001). Twenty-five mL of cells at an O.D.600 of 0.4–0.5 was fixed with 1% formaldehyde. For mock immunoprecipitation or beads alone (B), 25 µg of sepharose protein A beads per reaction were pre-incubated with 5 µg of BSA, 50 µg sonicated salmon sperm, and 500 µL of ChIP lysis buffer. For immunoprecipitations, 200 µL of 12CA5 culture supernatant (anti-HA immunoprecipitations) were incubated with sepharose protein A beads, BSA, and salmon sperm for 1 h at room temperature. For TAP tag immunoprecipitations, 20 µL of IgG sepharose slurry (Pharmacia) was used (Rpa190 and Rpa43 were TAP tagged). Cells were lysed in 100 µL of cold lysis buffer with 1×protease inhibitor cocktail (Complete Protease Inhibitors [Roche] in addition to Peflac, chymostatin, and benzamidine) by vortexing with glass beads for 3 min in the cold. The total volume of extract was brought to 750 µL and sonicated four times for 15 sec on ice. Aliquots were taken for input (5 µL or 1/50) and sheared DNA controls (100 µL). Cellular debris was pelleted, and 500 µL of extract was precleared with beads alone for 30 min at 4°C. After incubation for 1 h at 4°C with cell extract and antibody-bound beads, the beads were washed and eluted according to Rusche and Rine (2001). PCR amplification was carried out with 1 µL template, 40 ng of each primer, and Eppendorf MasterMix in a 20-µL total reaction. Input DNA was diluted 2.5-fold to ensure that the reaction occurred in the linear range. The oligonucleotides used were the following: 5′ETS rDNA 5′: 5′-GAATAGCCGGTCGCAAGACTG-3′ (+300 from Start) and 3′: 5′-ACCTATTCCCTCTTGCTAGAAG-3′ (+600 from Start). The DNA precipitate was denatured at 94°C for 2 min. The PCR product was amplified for 20 cycles of 94°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min with a final extension of 2 min at 72°C. After agarose gel electrophoresis, bands were quantified on an Alpha Innotech Imager. The value from beads alone (B) and anti-HA immunoprecipitation (IP) were normalized to total input for each ChIP. The beads-alone value was subtracted from the anti-HA immunoprecipitation value to obtain the indicated numbers.
Protein manipulations
When protein levels were examined, protein was extracted from early log-phase cells (O.D.600 0.4–0.5), and 10 µg of each sample, as determined using the Bradford method, was separated by SDS-PAGE. Coimmunoprecipitations were carried out as described by Wehner and Baserga (2002) except for coimmunoprecipitations with YJV100, in which twice as many cells, beads, and antibody was used to compensate for decreased protein levels. Gradients of 10%–47% and 10%–50% sucrose were carried out as described (Dragon et al. 2002). Western blots were carried out with antibodies described by Wehner et al. (2002) and 1:5000 anti-PAP (to recognize the TAP tag; Sigma).
Acknowledgments
We thank Laura Rusche and Jasper Rine for the ChIP protocol, Elsebet Lund for helpful suggestions, and Baserga lab members T. Paul Stone, Alicia Lee, Veni Manickam, Kara Bernstein, and Erica Champion for advice, discussion, and technical assistance. We thank Dick Raue and M. Nomura for the gift of yeast strains, and Joan Steitz and Peter Glazer for reading the manuscript. J.E.G.G. was supported by a pre-doctoral NRSA from the NIH, and S.G. was supported by Leslie H. Warner and Anna Fuller Fellowships in Cancer Research. This work was supported by NIHGMS grant 52581 to S.J.B. and NSF grant MCB-9513589 to A.L.B. S.J.B. is a member of the Yale Cancer Center.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1226604.
References
- Ausubel F., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. 1995. Short protocols in molecular biology. Wiley, New York.
- Bassler J., Grandi, P., Gadal, O., Lessmann, T., Petfalski, E., Tollervey, D., Lechner, J., and Hurt, E. 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell 8: 517-529. [DOI] [PubMed] [Google Scholar]
- Beltrame M. and Tollervey, D. 1992. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 11: 1531-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame M., Henry, Y., and Tollervey, D. 1994. Mutational analysis of an essential binding site for the U3 snoRNA in the 5′ external transcribed spacer of yeast pre-rRNA. Nucleic Acids Res. 22: 5139-5147. [Corrected and republished article originally printed in 1994 Nucleic Acids Res. 22: 4057-4065]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassiday L.A. and Maher III, L.J. 2002. Having it both ways: Transcription factors that bind DNA and RNA. Nucleic Acids Res. 30: 4118-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colley A., Beggs, J.D., Tollervey, D., and Lafontaine, D.L. 2000. Dhr1p, a putative DEAH-box RNA helicase, is associated with the box C+D snoRNP U3. Mol. Cell. Biol. 20: 7238-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver G.M. 2002. Sno-capped: 5′ ends of preribosomal RNAs are decorated with a U3 snoRNP. Chem. Biol. 9: 777-779. [DOI] [PubMed] [Google Scholar]
- Dragon F., Gallagher, J.E., Compagnone-Post, P.A., Mitchell, B.M., Porwancher, K.A., Wehner, K.A., Wormsley, S., Settlage, R.E., Shabanowitz, J., Osheim, Y., et al. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417: 967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar D.A., Wormsley, S., Agentis, T.M., and Baserga, S.J. 1997. Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol. Cell. Biol. 17: 5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E.A. and Warner, J.R. 1986. An RNA polymerase I enhancer in Saccharomyces cerevisiae. Mol. Cell. Biol. 6: 2089-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A., Cronshaw, A.D., Dlakic, M., and Tollervey, D. 2002. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell 9: 341-351. [DOI] [PubMed] [Google Scholar]
- Gavin A.C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J.M., Michon, A.M., Cruciat, C.M., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141-147. [DOI] [PubMed] [Google Scholar]
- Gerbi S.A., Borovjagin, A.V., Ezrokhi, M., and Lange, T.S. 2001. Ribosome biogenesis: Role of small nucleolar RNA in maturation of eukaryotic rRNA. Cold Spring Harbor Symposium Quant. Biol. 66: 575-590. [DOI] [PubMed] [Google Scholar]
- Gotta M., Strahl-Bolsinger, S., Renauld, H., Laroche, T., Kennedy, B.K., Grunstein, M., and Gasser, S.M. 1997. Localization of Sir2p: The nucleolus as a compartment for silent information regulators. EMBO J. 16: 3243-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P., Rybin, V., Bassler, J., Petfalski, E., Strauss, D., Marzioch, M., Schafer, T., Kuster, B., Tschochner, H., Tollervey, D., et al. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10: 105-115. [DOI] [PubMed] [Google Scholar]
- Granneman S. and Baserga, S.J. 2004. Ribosome biogenesis: Of knobs and RNA processing. Exp. Cell Res. 296: 43-50. [DOI] [PubMed] [Google Scholar]
- Harnpicharnchai P., Jakovljevic, J., Horsey, E., Miles, T., Roman, J., Rout, M., Meagher, D., Imai, B., Guo, Y., Brame, C.J., et al. 2001. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell 8: 505-515. [DOI] [PubMed] [Google Scholar]
- Ho Y., Gruhler, A., Heilbut, A., Bader, G.D., Moore, L., Adams, S.L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180-183. [DOI] [PubMed] [Google Scholar]
- Howe K.J. 2002. RNA polymerase II conducts a symphony of pre-mRNA processing activities. Biochim. Biophys. Acta 1577: 308-324. [DOI] [PubMed] [Google Scholar]
- Hughes J.M. 1996. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J. Mol. Biol. 259: 645-654. [DOI] [PubMed] [Google Scholar]
- Hughes J.M.X. and Ares Jr., M. 1991. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and prevents formation of 18S ribosomal RNA. EMBO J. 10: 4231-4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R., Tollervey, D., and Hurt, E.C. 1993. A U3 snoRNP protein with homology to splicing factor PRP4 and G β domains is required for ribosomal RNA processing. EMBO J. 12: 2549-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D., Linder, P., and de la Cruz, J. 1999. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 7897-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.J., Peng, W.T., Cagney, G., Robinson, M.D., Haw, R., Zhong, G., Guo, X., Zhang, X., Canadien, V., Richards, D.P., et al. 2004. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell 13: 225-239. [DOI] [PubMed] [Google Scholar]
- Lee S.J. and Baserga, S.J. 1997. Functional separation of pre-rRNA processing steps revealed by truncation of the U3 small nucleolar ribonucleoprotein component, Mpp10. Proc. Natl. Acad. Sci. 94: 13536-13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit P., Kuhn, H., Gas, N., and Tschochner, H. 2003a. The pre-ribosomal network. Nucleic Acids Res. 31: 799-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit P., Strauss, D., Bassler, J., Gadal, O., Kuhn, H., Schutz, S., Gas, N., Lechner, J., Hurt, E., and Tschochner, H. 2003b. A Noc complex specifically involved in the formation and nuclear export of ribosomal 40 S subunits. J. Biol. Chem. 278: 4072-4081. [DOI] [PubMed] [Google Scholar]
- Nissan T.A., Bassler, J., Petfalski, E., Tollervey, D., and Hurt, E. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21: 5539-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Yano, R., and Nomura, M. 1991. Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc. Natl. Acad. Sci. 88: 3962-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim Y.N. and Beyer, A.L. 1989. Electron microscopy of RNP complexes on nascent RNA using the Miller chromatin spreading method. Methods Enzymol. 180: 481-509. [DOI] [PubMed] [Google Scholar]
- Peculis B.A. 2002. Ribosome biogenesis: Ribosomal RNA synthesis as a package deal. Curr. Biol. 12: R623-R624. [DOI] [PubMed] [Google Scholar]
- Peng W.T., Robinson, M.D., Mnaimneh, S., Krogan, N.J., Cagney, G., Morris, Q., Davierwala, A.P., Grigull, J., Yang, X., Zhang, W., et al. 2003. A panoramic view of yeast noncoding RNA processing. Cell 113: 919-933. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. and O'Sullivan, J. 2002. Polyadenylation: A tail of two complexes. Curr. Biol. 12: R855-R857. [DOI] [PubMed] [Google Scholar]
- Roger B., Moisand, A., Amalric, F., and Bouvet, P. 2002. Repression of RNA polymerase I transcription by nucleolin is independent of the RNA sequence that is transcribed. J. Biol. Chem. 277: 10209-10219. [DOI] [PubMed] [Google Scholar]
- ____. 2003. Nucleolin provides a link between RNA polymerase I transcription and pre-ribosome assembly. Chromosoma 111: 399-407. [DOI] [PubMed] [Google Scholar]
- Rusche L.N. and Rine, J. 2001. Conversion of a gene-specific repressor to a regional silencer. Genes & Dev. 15: 955-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarsky D.A. and Fournier, M.J. 1998. Functional mapping of the U3 small nucleolar RNA from the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 3431-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveanu C., Namane, A., Gleizes, P.E., Lebreton, A., Rousselle, J.C., Noaillac-Depeyre, J., Gas, N., Jacquier, A., and Fromont-Racine, M. 2003. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 23: 4449-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer T., Strauss, D., Petfalski, E., Tollervey, D., and Hurt, E. 2003. The path from nucleolar 90S to cytoplasmic 40S preribosomes. EMBO J. 22: 1370-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K. and Tollervey, D. 1999. Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol. Cell. Biol. 19: 6012-6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K., Kawasaki, Y., Hiraga, S., Tawaramoto, M., Nakashima, N., and Sugino, A. 2002. The fifth essential DNA polymerase phi in Saccharomyces cerevisiae is localized to the nucleolus and plays an important role in synthesis of rRNA. Proc. Natl. Acad. Sci. 99: 9133-9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou W., Sakamoto, K.M., Keener, J., Morimoto, K.W., Traverso, E.E., Azzam, R., Hoppe, G.J., Feldman, R.M., De-Modena, J., Moazed, D., et al. 2001. Net1 stimulates RNA polymerase I transcription and regulates nucleolar structure independently of controlling mitotic exit. Mol. Cell 8: 45-55. [DOI] [PubMed] [Google Scholar]
- Strasser K., Masuda, S., Mason, P., Pfannstiel, J., Oppizzi, M., Rodriguez-Navarro, S., Rondon, A.G., Aguilera, A., Struhl, K., Reed, R., et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304-308. [DOI] [PubMed] [Google Scholar]
- Torchet C. and Hermann-Le Denmat, S. 2000. Bypassing the rRNA processing endonucleolytic cleavage at site A2 in Saccharomyces cerevisiae. RNA 6: 1498-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschochner H. and Hurt, E. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends. Cell Biol. 13: 255-263. [DOI] [PubMed] [Google Scholar]
- Venema J. and Tollervey, D. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33: 261-311. [DOI] [PubMed] [Google Scholar]
- Venema J., Dirks-Mulder, A., Faber, A.W., and Raue, H.A. 1995. Development and application of an in vivo system to study yeast ribosomal RNA biogenesis and function. Yeast 11: 145-156. [DOI] [PubMed] [Google Scholar]
- Warner J.R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24: 437-439. [DOI] [PubMed] [Google Scholar]
- ____. 2001. Nascent ribosomes. Cell 107: 133-136. [DOI] [PubMed] [Google Scholar]
- Watkins N.J., Segault, V., Charpentier, B., Nottrot, S., Fabrizio, P., Bachi, A., Wilm, M., Rosbash, M., Branlant, C., and Luhrmann, R. 2000. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103: 457-466. [DOI] [PubMed] [Google Scholar]
- Wehner K.A. and Baserga, S.J. 2002. The σ70-like motif: A eukaryotic RNA binding domain unique to a superfamily of proteins required for ribosome biogenesis. Mol. Cell 9: 329-339. [DOI] [PubMed] [Google Scholar]
- Wehner K.A., Gallagher, J.E., and Baserga, S.J. 2002. Components of an interdependent unit within the SSU processome regulate and mediate its activity. Mol. Cell. Biol. 22: 7258-7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston K. and Bishop, J.M. 1989. Transcriptional activation by the v-myb oncogene and its cellular progenitor, c-myb. Cell 58: 85-93. [DOI] [PubMed] [Google Scholar]
- Wu L.F., Hughes, T.R., Davierwala, A.P., Robinson, M.D., Stoughton, R., and Altschuler, S.J. 2002. Large-scale prediction of Saccharomyces cerevisiae gene function using overlapping transcriptional clusters. Nat. Gen. 31: 255-265. [DOI] [PubMed] [Google Scholar]