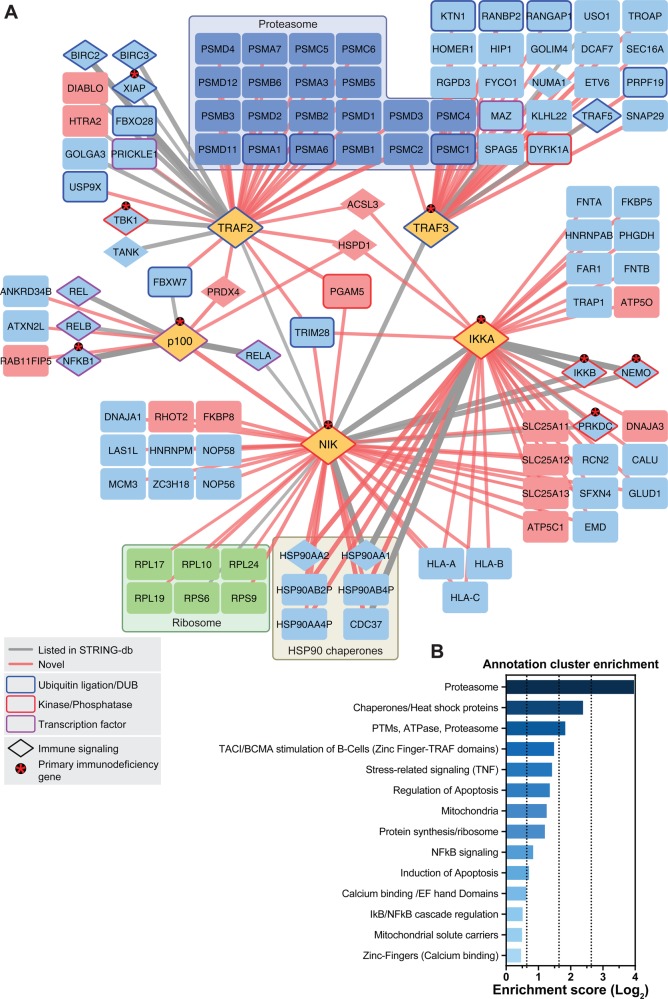
Figure 2.
Interaction network of the noncanonical NF-κB signaling pathway. (A) Proteins shown were identified by LC–MS. Edge (line) thickness and edge transparency represent the CRAPome FC-A score. Red edges indicate novel interactions, and gray edges indicate known interactions. SH-tagged proteins, proteasomal machinery proteins, ribosomal proteins, and mitochondrial-localizing proteins are colored yellow, dark-blue, green, and red, respectively. Ubiquitination/deubiquitination machinery proteins, kinases/phosphatases, and transcription factors are framed in blue, red, and purple, respectively. Asterisks denote proteins that cause primary immunodeficiency phenotypes in humans when mutated. (B) Gene Ontology term enrichment analysis using DAVID functional annotation clustering. Selected clusters are shown.