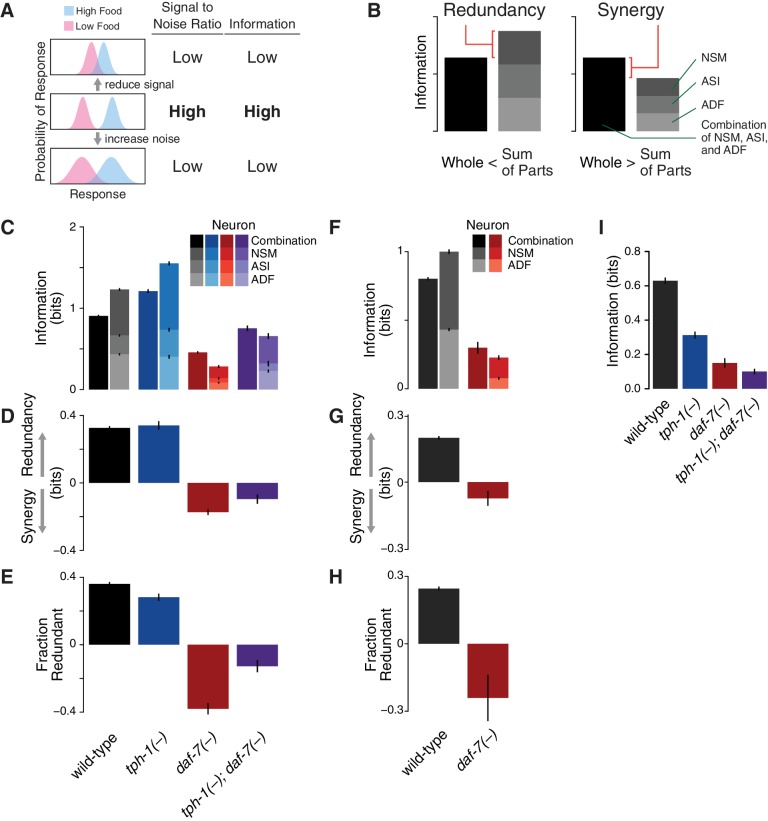
Figure 1. Redundancy and synergy in a gene expression code.
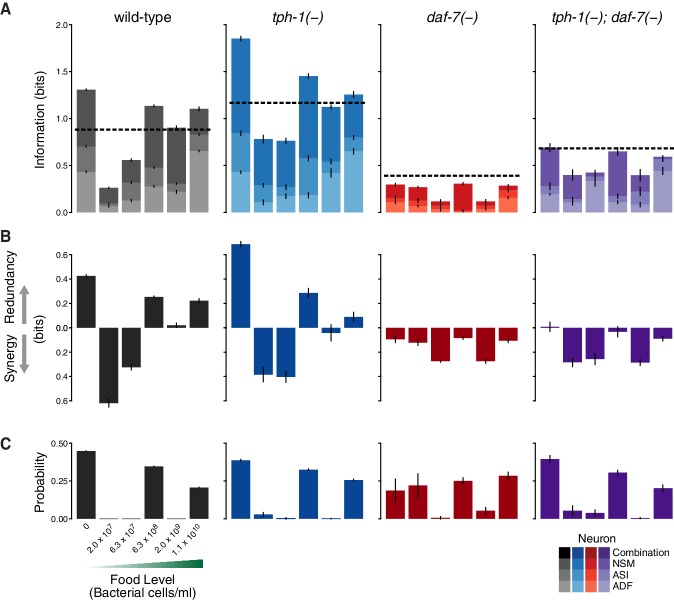
(A) Information content depends on the overlap between gene expression distributions under different environmental conditions, which in turn depends on both the response magnitude (signal) and the variability across the population (noise). (B) Diagrams illustrating redundancy versus synergy, calculated as the difference between the whole (combinatorial information in NSM/ASI/ADF; darkest bar) and the sum of parts (information in NSM + ASI + ADF; stacked bars). (C–E) Analysis of redundancy and synergy based on tph-1 expression in ADF and NSM, and daf-7 expression in ASI. Genotype color key: Wild-type (black), tph-1(-) (blue), daf-7(-) (red), and tph-1(-); daf-7(-) (purple). (C) Effect of tph-1(-) and daf-7(-) mutations on food encoding in the whole circuit (darkest bars) and the sum of parts (lighter stacked bars). (D) Effect of tph-1(-) and daf-7(-) on redundancy and synergy among ADF, NSM, and ASI, as defined in Equation 2 and (B). As described in Equation 2 and in the main text, redundancy and synergy are indicated by positive and negative R values, respectively. (E) Fraction of redundant or synergistic information in ADF, NSM, and ASI, which is the amount of redundancy or synergy in (D) normalized to the information encoded. (F–H) Analysis of redundancy and synergy only in the tph-1 expressing neurons, ADF, and NSM. (F) Effect of daf-7(-) in the information encoded by tph-1 expression in ADF and NSM (darkest bars) and the sum of their parts (lighter stacked bars). (G) Effect of daf-7(-) on redundancy/synergy of ADF and NSM. (H) Fraction of redundant or synergistic information in tph-1 expression in ADF and NSM, which is the amount of redundancy or synergy in (G) normalized to the total information encoded from (F). (I) Loss of tph-1 and daf-7 degrades information about food abundance at the level of lifespan responses.
DOI: http://dx.doi.org/10.7554/eLife.24040.002
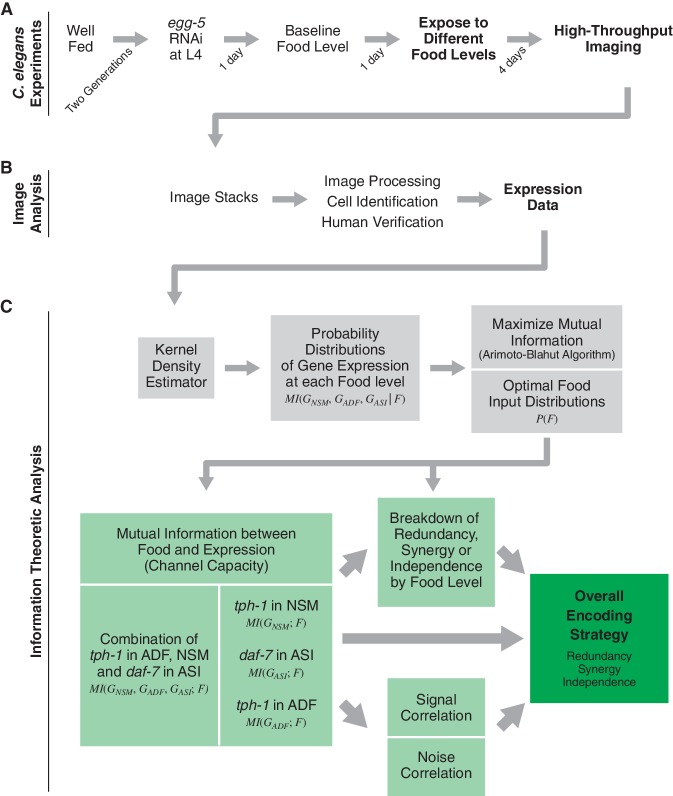
Figure 1—figure supplement 1. Schematic of experimental and analytical workflow.
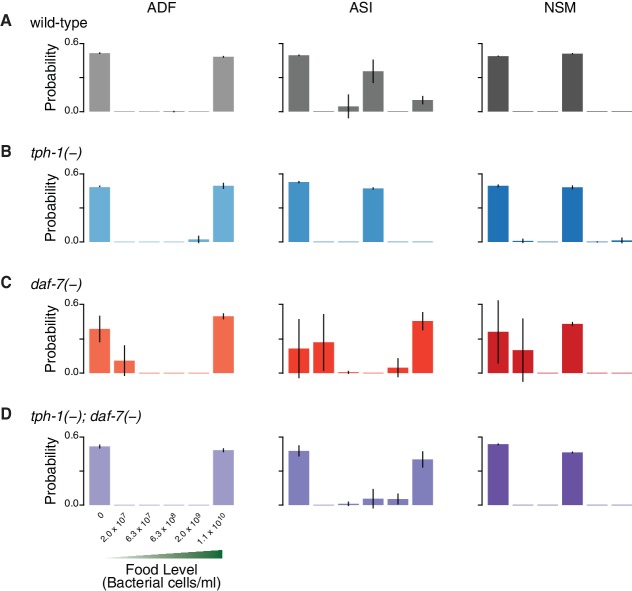
Figure 1—figure supplement 2. Experimental variability.