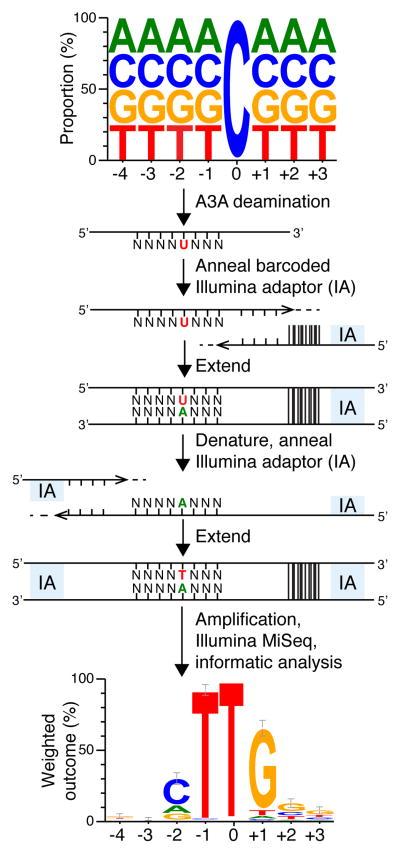
Figure 1. Deep-deamination approach to determine an optimal human A3A substrate.
A ssDNA library with a single target C and N’s on the 5′ and 3′ sides was reacted with human A3A (near-single hit kinetics). The resulting pool containing C-to-U deamination products was annealed to a bar-coded Illumina adaptor (IA) and T4 DNA polymerase was used to produce a complementary DNA strand. This intermediate was denatured, annealed to a 5′-IA, and converted to duplex by Phusion thermostable high fidelity DNA polymerase. Illumina Mi-Seq was used to generate reads for subsequent informatics analysis. A weblogo representation of deamination products unique to A3A shows enrichment for −1 T and +1 G that informed the ssDNA sequence for co-crystallization experiments (n=641; error bars are twice the sample correction value). Source data are provided as a spreadsheet (.xlsx) linked to the online legend.