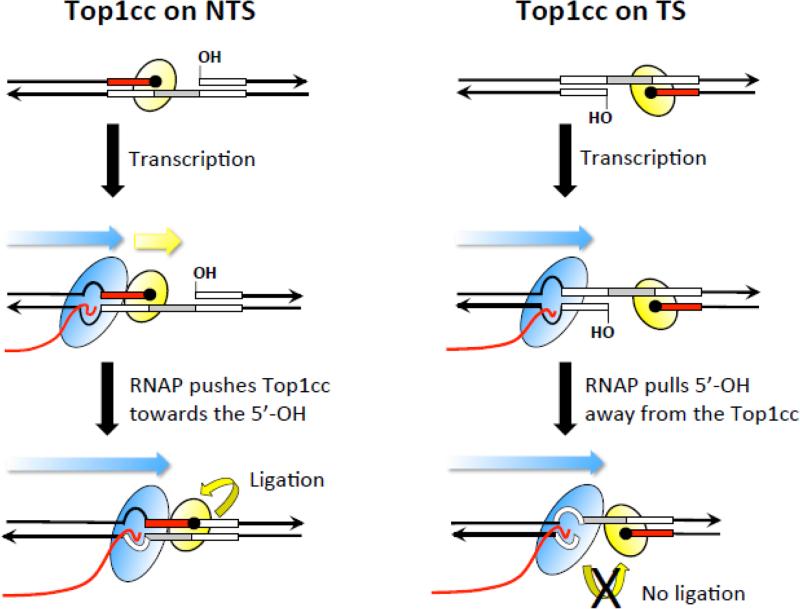
Figure 4.
Encounters with RNAP promote deletions only when Top1 cleaves the NTS. Black lines correspond to DNA and arrowheads indicate 3’ ends. A deletion hotspot with three repeat units (boxes) is shown in each panel; inversion of the hotspot moves the cleavage sites from the NTS to the TS in the panels on the left and right, respectively. The Top1cc (yellow oval) is trapped at the end of the red repeat; the repeat within the gap created by sequential Top1 cleavage is in gray. The 5’-OH that borders the gap was generated by the first Top1 incision at a ribonucleotide. The blue arrow depicts the direction of RNAP (blue oval) movement and the trailing red line is the nascent RNA. When the Top1cc is trapped on the NTS, the approaching RNAP pushes it (yellow arrow) and the attached repeat towards the 5’-OH to facilitate ligation. When the Top1cc is trapped on the TS, an advancing RNAP pulls the 5’-OH away from the Top1cc, preventing Top1-mediated ligation.