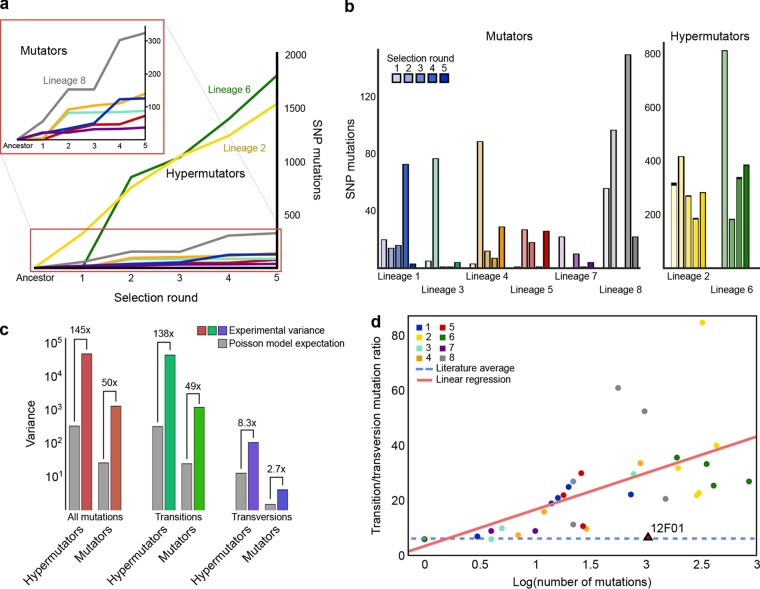
FIG 1 .
Serial selection for salt tolerance identifies a hypermutator phenotype, with a distinct mutation profile, in Vibrio splendidus 12B01. (a) We grew eight independent lineages on hypersaline media and sequenced genomes from each selection round. Strict SNP calling indicated that two hypermutator lineages had rapidly accumulated a large number of mutations, but all lineages had accumulated many more mutations (37 to 1,802) than the one or two mutations expected, given literature averages of spontaneous mutation (2). (b) The number of new mutations varied greatly across selection rounds in both mutator and hypermutator lineages. (c) This variability was much higher than the expected Poisson distribution variance, and this disparity was largely driven by transition mutations. (d) Selection rounds with more mutations tended to have larger ratios of transition versus transversion mutations; these ratios far exceeded averages from the literature (28), indicated by a dashed blue line, and the transition-to-transversion ratio of 12B01 compared with a closely related strain, V. splendidus 12F01.