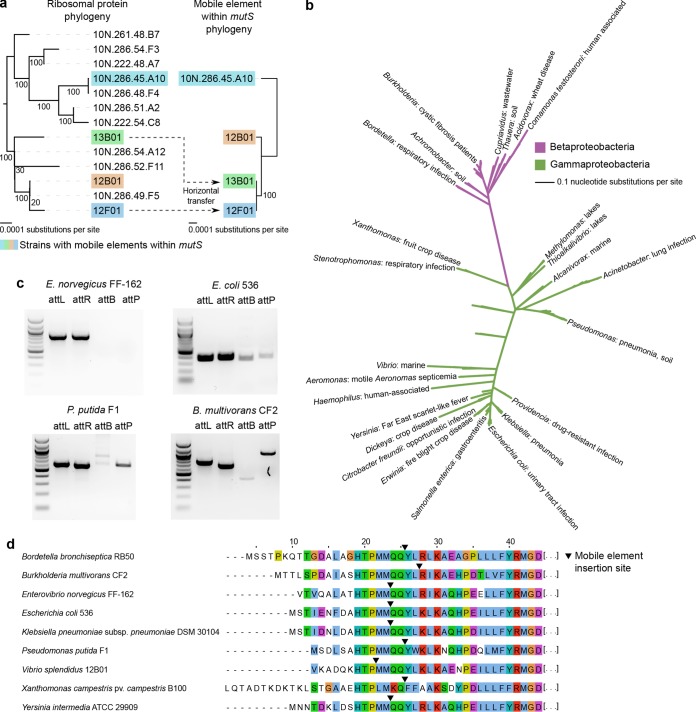
FIG 3 .
Mobile elements within mutS occur across Vibrio, Betaproteobacteria, and Gammaproteobacteria. (a) Phylogeny of close relatives of 12B01 (>98% similarity in 16S rRNA). Strains with mobile elements within mutS are not a monophyletic clade, and comparison between host phylogenies and mobile element phylogenies indicates that these elements have been horizontally transferred (dotted lines). (b) Broader BLAST searches identified other bacteria with mobile elements within mutS, including many human pathogens (Table S2). Phylogenetic tree built using 16S rRNA sequences. (c) Using a PCR assay similar to what we used for 12B01, we found mobile element excision in some, but not all, of a small subset of these bacteria when grown to the stationary phase. Sanger sequencing of the attB and attP PCR products from E. coli 536, P. putida F1, and B. multivorans CF2 indicated that mobile element excision in these bacteria did not result in any deletions or frameshift mutations. Expected amplicon lengths: E. norvegicus FF-162 attL = 894 bp, attR = 891 bp, attB (hypothetical) = 380 bp, and attP (hypothetical) = 1,405 bp; E. coli 536 attL = 404 bp, attR = 416 bp, attB = 393 bp, and attP = 427 bp; P. putida F1 attL = 601 bp, attR = 583 bp, attB = 596 bp, and attP = 588 bp; and B. multivorans CF2 attL = 811 bp, attR = 723 bp, attB = 451 bp, and attP = 1,083 bp. (d) These mobile elements were all integrated into the HTPMMQQ amino acid motif in MutS, although the precise location varied.