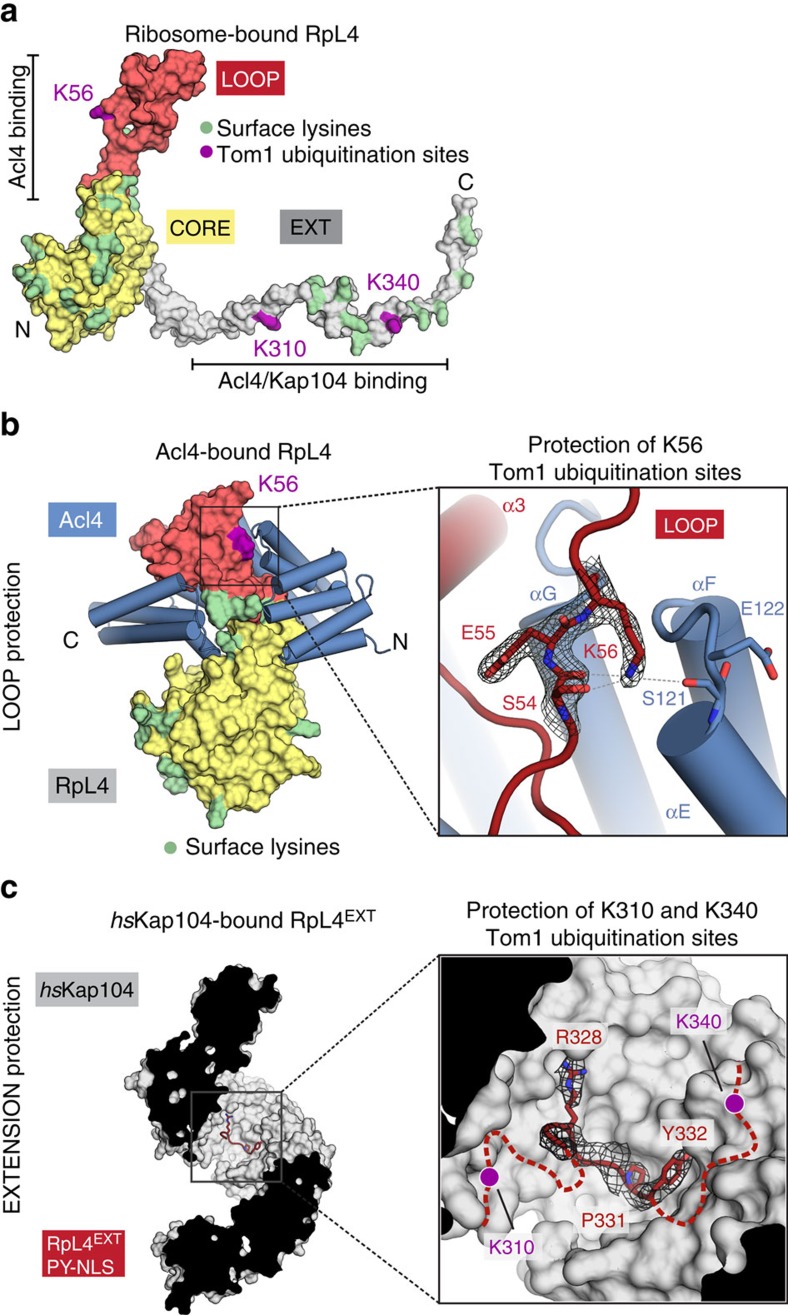
Figure 4. Shielding of Tom1 ubiquitination sites.
(a) Surface representation of ribosome-bound RpL4 (PDB ID 4V88)17. Acl4 and Kap104 binding sites are indicated with black bars. (b) Acl4-bound RpL4 (coloured as in Fig. 1a) and Acl4 (blue) are shown in surface and cartoon representation, respectively. The inset marks the Tom1 ubiquitination site that is illustrated in detail on the right. Acl4 (blue) and RpL4 (red) and critical residues highlighted in stick representation with a section of the final 2|Fo|-|Fc| electron density map contoured at 1.0 σ. (c) Crystal structure of the hsKap104·RpL4EXT complex. The inset marks the location of the Kap104 PY-NLS binding site that is illustrated in detail on the right. The residues of the PY-NLS consensus sequence, Arg328, Pro331 and Tyr332 are highlighted in stick representation with a section of the final 2|Fo|-|Fc| electron density map contoured at 1.0 σ. Magenta circles indicate the approximate location of RpL4 residues K310 and K340 that are ubiquitinated by Tom1 in the absence of Kap104.