Figure 6. Length distribution of string-like collective rearrangements of the atoms within ubiquitin molecule in glycerol, water, and a glycerol-water mixture at T = 300 K.
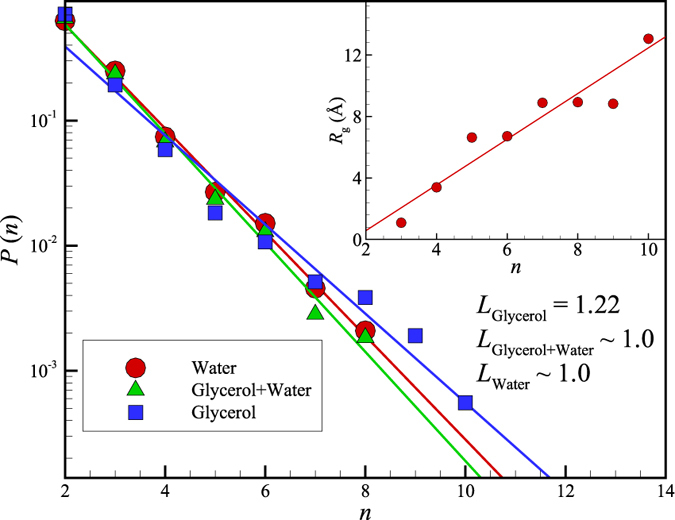
The inset displays the average radius of gyration of the ubiquitin strings, Rg, as a function of the average string length L = 〈n〉 in glycerol, the only case where collective motion is appreciable at this T in our CHARMM36 simulations. We see that L is near 1 in both the pure water and water/glycerol mixture, indicating that the TIP3P water has essentially suppressed all collective exchange motion within the protein, proving further evidence that the protein dynamics is ‘slaved’ to the dynamics of the solvent. The average radius of gyration Rg describes the average spatial extent of the dynamic clusters, while L describes their average contour length. Note that the strings are highly polydisperse, which tends to make the average values of Rg and L rather small in comparison to larger strings seen in a visualization of collective atomic exchange events within the protein.
