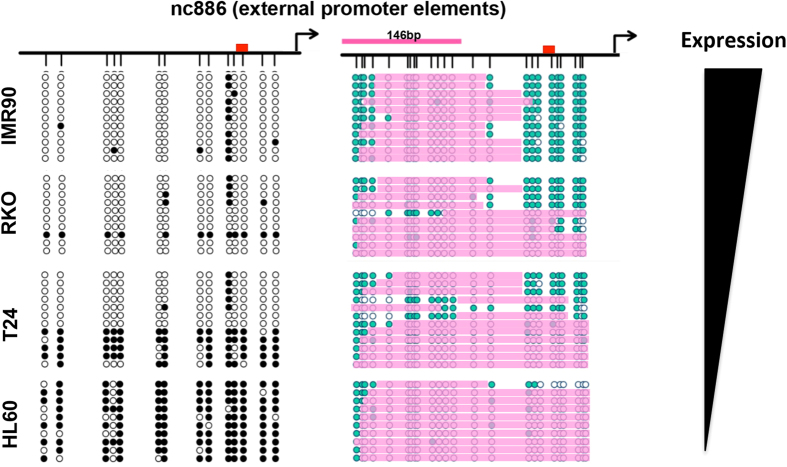
Figure 5. Accessibility mapping of the promoter structure at nc886.
NOMe-seq at the nc886 promoter in RKO, IMR90, HL60 and T24 cells. Left panel indicates endogenous DNA methylation, right panel chromatin accessibility. Each horizontal row in each panel represents the same DNA molecule, and it is thus possible to track accessibility and DNA methylation patterns in the same DNA molecule. For the accessibility plots, teal circles represent accessible GpC sites, white inaccessible. If regions of consecutive inaccessible sites equal or exceed 146 bp, we indicate the region as being occupied by a nucleosome (pink bars). For the DNA methylation plots, white indicates unmethylated, black methylated. Red box represents the CRE-element. Relative expression of nc886 between the four cell lines is indicated in the outer right panel.