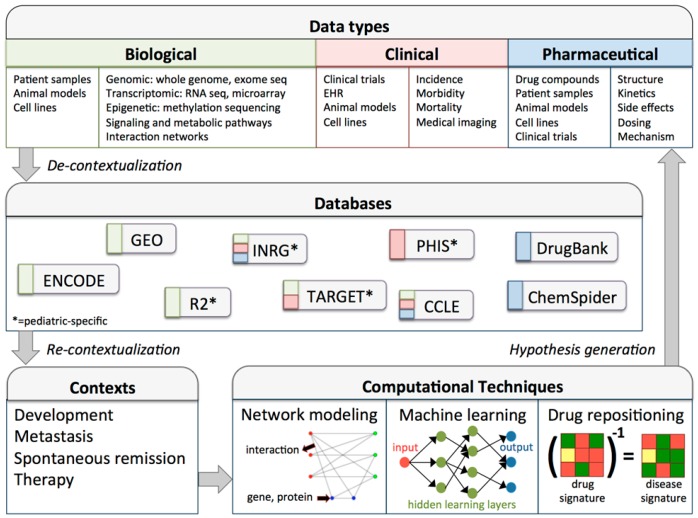
Figure 1.
Data-driven research workflow for neuroblastoma or other pediatric cancers. The data types panel enumerates data sources (e.g., animal models) and types (e.g., RNA sequencing) for three categories of biological data. Following collection, data may be de-contextualized for inclusion in databases. De-contextualization involves properly formatting and annotating datasets, so that they are standardized, accessible, and useful for re-contextualization into new research contexts. Note that this workflow is iterative, and can therefore benefit from continued improvement of data infrastructure and data collections, development of more accurate and comprehensive data analysis tools, and advances in basic and translational research and therapeutic applications [35,118,119,120,121,122,123,124]. CCLE, cancer cell line encyclopedia; ENCODE, encyclopedia of DNA elements; GEO, gene expression omnibus; EHR: electronic health record; INRG, international neuroblastoma risk group; PHIS, pediatric health information system; R2, genomics analysis and visualization platform; TARGET, therapeutically applicable research to generate effective treatments.