Figure 1.
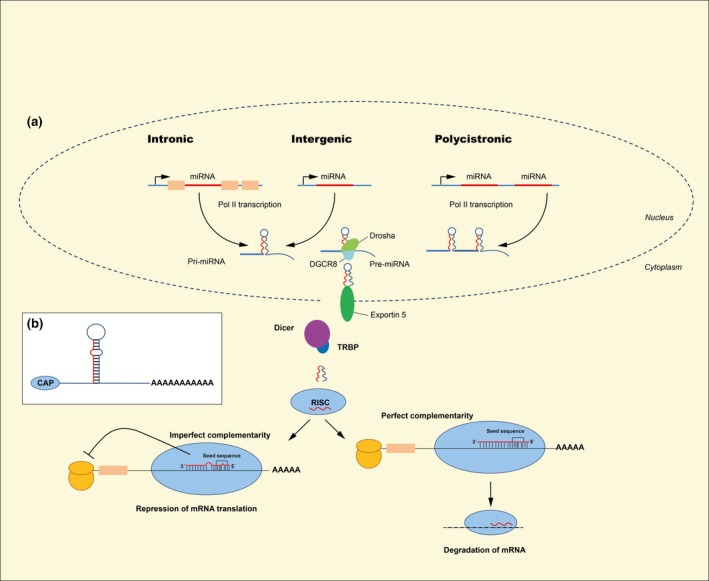
The canonical miRNA biogenesis pathway (a) and the average precursor miRNA (b). (a): The miRNA genes lies either intronic, intergenic or polycistronic. The primary miRNA (Pri‐miRNA) is transcribed by polymerase II (or polymerase III). The Pri‐miRNA is cleaved by the microprocessor complex Drosha‐DGCR8 in the nucleus. The precursor miRNA (pre‐miRNA) is transported out in the cytoplasm by Exportin 5. In the cytoplasm, the pre‐miRNA is further cleaved to its mature length (approx. 22 nt) by the RNase III Dicer in complex with the double‐stranded RNA‐binding protein TRBP. Argonaute (AGO2) proteins unwind the miRNA duplex and facilitate incorporation of the guide strand (red) into the RNA‐induced silencing complex (RISC). AGO2 then guides the RISC miRNA assembly to target mRNAs, whereas the passenger strand (blue) is degraded. Some miRNA bind mRNA with perfect complementarity and induce degradation of mRNA. miRNA also bind to targets with imperfect complementarity and block translation. (b): An average precursor miRNA with a hairpin stem of 33 base pairs, a terminal loop and two flanking region, where 5′ end are capped and a polyadenylated 3′ end.
