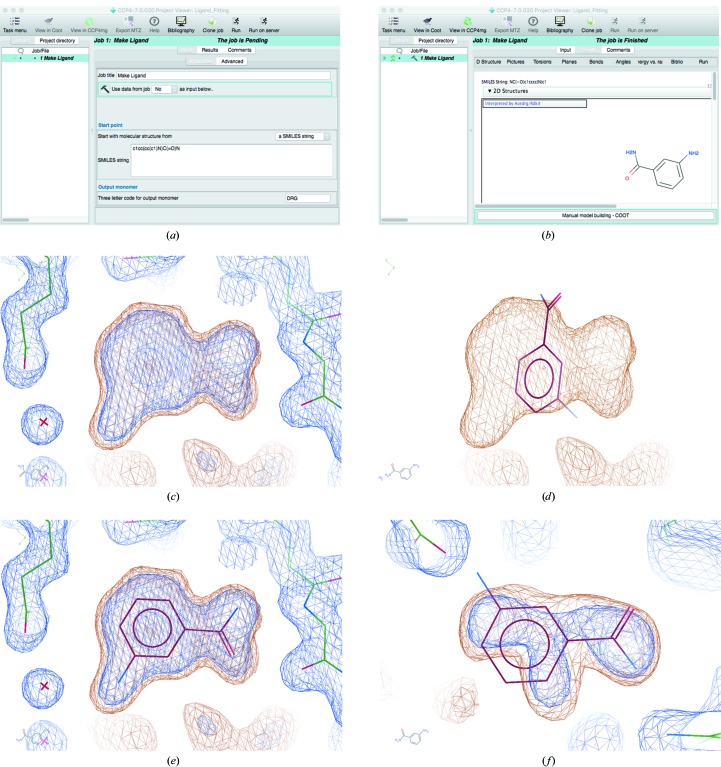
Figure 2.
Automatic ligand fitting in CCP4. The SMILES string corresponding to 3-aminobenzamide is pasted into the ‘Make Ligand’ task interface in CCP4i2 (a). Upon running the job, AceDRG is used to generate ligand restraints and RDKit is used to generate an initial conformer and a two-dimensional representation of the ligand (b). The ‘Manual Model Building’ task is then executed to open Coot. (c) displays the model (sticks) and 2mF o − DF c density map (blue) corresponding to a structure solved with data extending to 2 Å resolution (PDB entry 3kcz; Karlberg et al., 2010 ▸) after manually removing the ligands. The maps are shown using Coot’s default contour levels. Automatic ligand fitting is performed, using 3-aminobenzamide as the target. The focal region corresponds to the top-ranked blob identified in the masked map (orange). The ligand coordinates are nominally positioned onto the centre of the blob (d). The ligand is then optimally oriented and rigid-body refined into the masked density (e). In this case, manual intervention would be required in order to ensure favourable hydrogen bonds are satisfied: this issue is further addressed in §6 and Fig. 7 ▸. Multiple blobs are found in the map and ligands are fitted into them; the third-highest ranked ligand is automatically fitted into a blob that actually corresponds to a glycerol molecule (f).