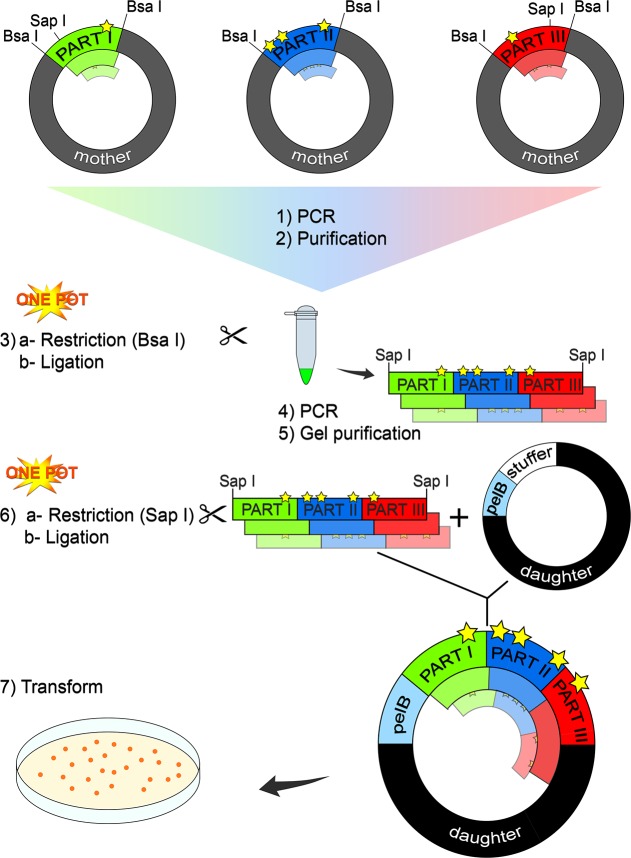
Fig 2. Facile reassembly of individually mutated gene parts.
The Cal-A gene was obtained as three separate parts in DNA2.0 mother vectors. In this method, the parts can be mutated independently as appropriate for each part (Table 1, yellow stars represent illustrative mutations). As a proof of concept to demonstrate the versatility of the method, NDT libraries were generated for parts 1 and 3, and part 2 was randomly mutated. The parts (both mutated and wild-type) were then amplified by PCR reactions. They were then purified (steps 1, 2 and S3 Fig) for assembly into a number among the possible combinations of mutated parts (see Table 1 for chosen combinations), in a one-pot restriction-ligation reaction using BsaI (3). The library of assembled genes was PCR amplified and gel purified (4, 5 and S4 Fig). Each amplified library was inserted into the daughter vector (6) using SapI in a one-pot restriction-ligation reaction, for transformation into E. coli (7). Note that a simplified version of this strategy is also possible, but was found to work only when applied to the wild-type parts (S1 and S5 Figs).