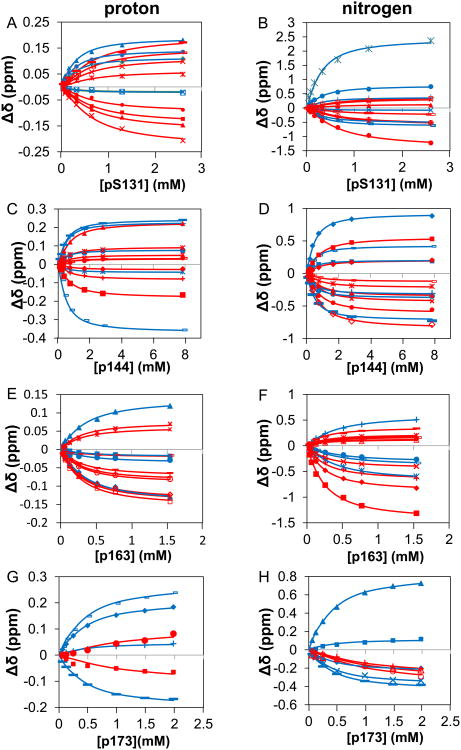
Figure 6. The simultaneous fits of intact Pin1 with singly phosphorylated IRAK1-UD derived peptides shown in terms of chemical shift perturbation of selected residues in individual dimensions.
The x-axis is the concentration of peptide titrated in and the y-axis is the chemical shift perturbation. Blue represents WW domain residues while red represents PPIase residues. The symbols are the experimental data and the lines are the simultaneous fits. A) selected residues in the proton dimension (F25 unfilled□, Q33 filled◊, W34 filled ○, W34e ×, E35 filledΔ, H59 filled○, R69 ×, S114 -, S115 *, R127 ×, G128 filledΔ, Q129 filled□, and M130 —) for Pin1 titrated with p131. B) selected residues in the nitrogen dimension (Y23 filledΔ, Y24 ×, Q33 *, W34 filled ○, W34e +, E35 -, V62 —, R69 filled◊, S114 unfilled□, A116 filledΔ, R127 ×, G128 unfilled ○, Q129 filled ○, M130 +) for Pin1 titrated with p131. C) selected residues in the proton dimension (Y24 *, F25 filled ○, W34 +, W34e -, E35 —, H59 filled◊, V62 -, R69 filled□, S114 filledΔ, S115 *, A116 ×, G128 +, Q129 filled ○) for Pin1 titrated with p144. D) selected residues in the nitrogen dimension (Y23 filled ○, Y24 +, F25 -, Q33 —, W34 filled◊, W34e filled□, V62 *, R69 unfilled◊, S114 filled□, S115 ×, A116 filled◊, R127 -, G128 filledΔ, Q129 filled ○) for Pin1 titrated with p144. E) selected residues in the proton dimension (Y23 filled□, Y24 filled ○, W34 filledΔ, W34e filled◊, H59 unfilledΔ, L61 ×, V62 *, R69 unfilled□, A116 unfilled ○, K117 —, A118 unfilled◊, G128 -, Q129 +) for Pin1 titrated with p163. F) selected residues in the nitrogen dimension (Y23 *, Y24 filled ○, W34 filled◊, E35 -, H59 —, V62 -, R69 filled◊, S115 filled□, A116 unfilledΔ, A118 ×, R127 filled ○, Q129 filled◊, K132 *) for Pin1 titrated with p163. G) selected residues in the proton dimension (F25 +, W34 -, W34e —, E35 filled◊, S114 filled□, S115 filled ○) for Pin1 titrated with p173 H) selected residues in the nitrogen dimension (Y23 —, Y24 filled◊, F25 filled□, W34 filledΔ, E35 ×, R68 unfilledΔ, Q129 unfilled ○, M130 +) for Pin1 titrated with p173.