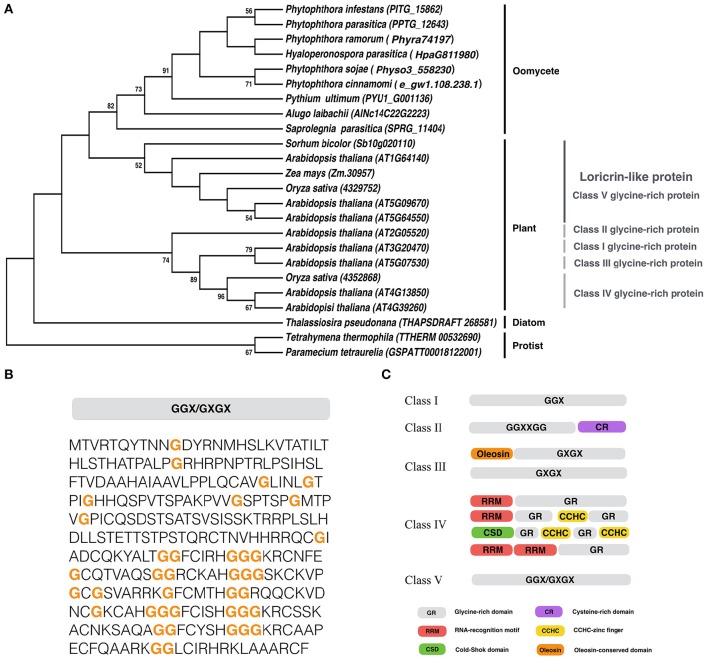
Figure 1.
Sequence analysis of PiLLP. (A) Phylogenetic analysis of LLP from different organisms. A maximum-likelihood tree was constructed using MEGA 5 with LLP sequences from the species including, Phytophthora cinannmomi, Phytophthora infestans, Phytophthora ramorum, Phytophthora sojae, Phytophthora parasitica, Albugo laibachii, Hyaloperonospora parasitica, Pythium ultimum, Saprolegnia parasitica, Thalassiosira pseudonana, Micromonas pusilla, Arabidopsis thaliana, Oryza sativa, Sorhum bicolor, Zea mays, Paramecium tetraurella, and Tetrahymena thermophile. Phylogenetic trees were constructed with the maximum likelihood evolution algorithm in MEGA 5.22. The statistical strengths were assessed by bootstraps with 1000 replicates. Bootstrap values (≥50%) are shown near the tree nodes. (B) The complete amino acid sequence of PiLLP (accession number XP_002898099). Orange bold letter, glycine repeat region. (C) Schematic representation of plant glycine-rich proteins classification. CCHC, CCHC-zinc finger; CSD, Cold-shock domain; CR, cysteine-rich domain; GR, Glycine rich domain; Oleosin, Oleosin-conserved domain; RRM, RNA-recognition motif. Glycine-rich repeats are indicated as GGX, GGXXXGG, GXGX, and GGX/GXGX, where G represents glycine and X represents any amino acid (Modified from Mangeon et al., 2010).