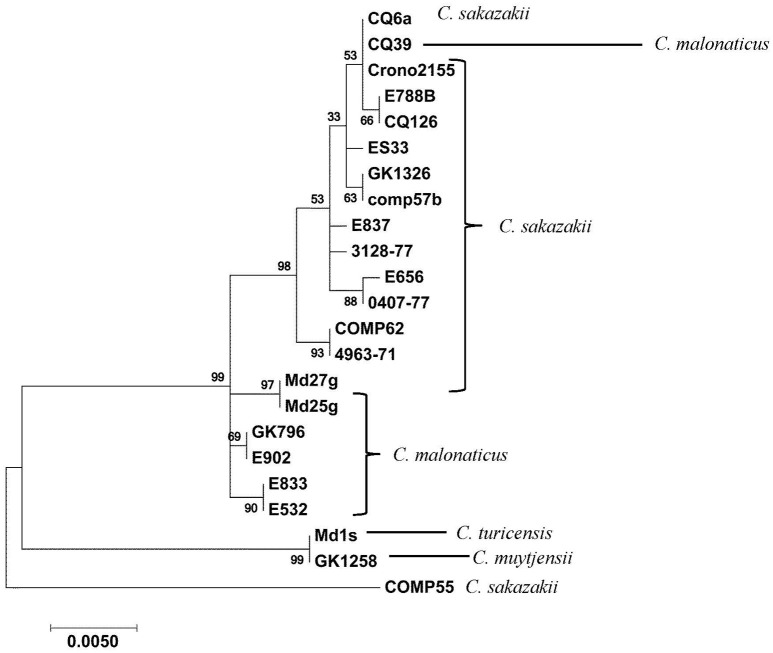
Figure 4.
ompA-gene based clustering of Cronobacter isolates highlight intra- and inter-species sequence diversity. Twenty-one alleles of ompA representing the 77 genomes analyzed in this work were aligned using ClustalW and clustered in the Maximum-likelihood method (Tamura and Nei, 1993) using the tools available on MEGA7 phylogenetic suite (Kumar et al., 2016). One thousand forty-four positions were evaluated for allelic information across the span of ompA gene sequences for the 21 strains. The bootstrap consensus (values located on the nodes) in the tree are inferred from 500 replicates. These values are taken to represent the evolutionary history of the taxa analyzed using the option available in MEGA7. The manual annotation of the tree highlights the intraspecies differences in C. sakazakii, C. turicensis, C. muytjensii, and C. malonaticus. The other members of the individual groups are listed in Supplemental Table 1 and those strains listed in the table and are identified by an asterisk (**) indicate those strains which had their ompA nucleotide sequences translated. Additionally those strains listed in this table, identified by a single (*), represent the strains included in this figure.