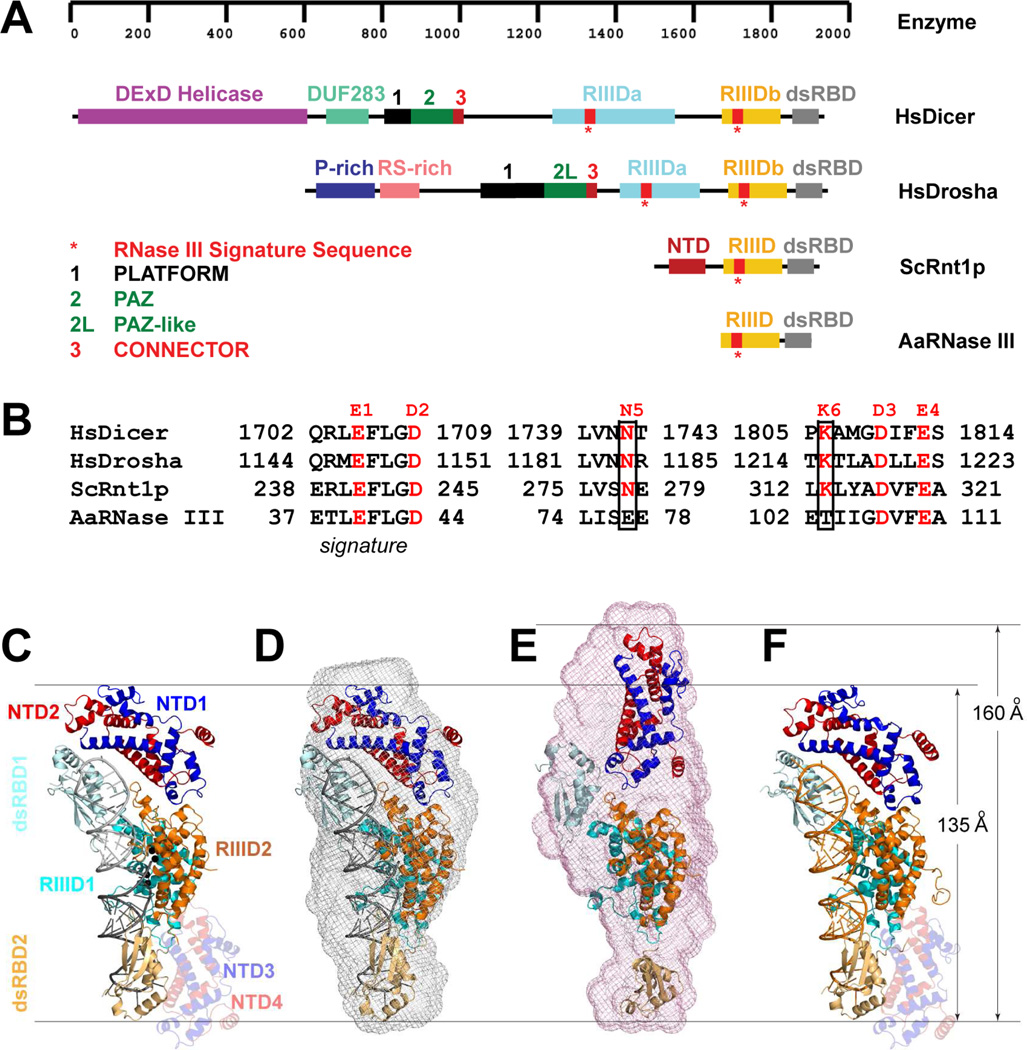
Figure 1.
Representative members of the RNase III family and the scope of this study. (A) Domain structures of Homo sapiens Dicer (HsDicer) (Kwon et al., 2016), Homo sapiens Drosha (HsDrosha) (Kwon et al., 2016), Saccharomyces cerevisiae Rnt1p (ScRnt1p) (Liang et al., 2014), and Aquifex aeolicus RNase III (AaRNase III) (Gan et al., 2006). The protein domains are color coded as indicated. (B) Catalytic amino acid residues in the RIIID of HsDicer, HsDrosha, ScRnt1p, and AaRNase III. The N5 and K6, unique for eukaryotes, are boxed. (C) Crystal structure of the Rnt1p post-cleavage complex (PDB entry 4OOG). The structure is illustrated as a ribbon diagram (helices as spirals, strands as arrows, loops as tubes, RNAs as tube-and-stick models, and Mg2+ ions as spheres). The protein domains are color coded. The extra NTD dimer (NTD3/NTD4) is shaded. (D) Crystal structure (PDB entry 4OOG, excluding the extra NTD dimer) docked into the SAXS envelope (this work). (E) Best-fit rigid-body model of apo-Rnt1p (starting structure: PDB entry 4OOG) docked into the SAXS envelope (this work). (F) Crystal structure of Rnt1p in complex with an RNA substrate analog, i.e., the substrate-loaded complex (this work).