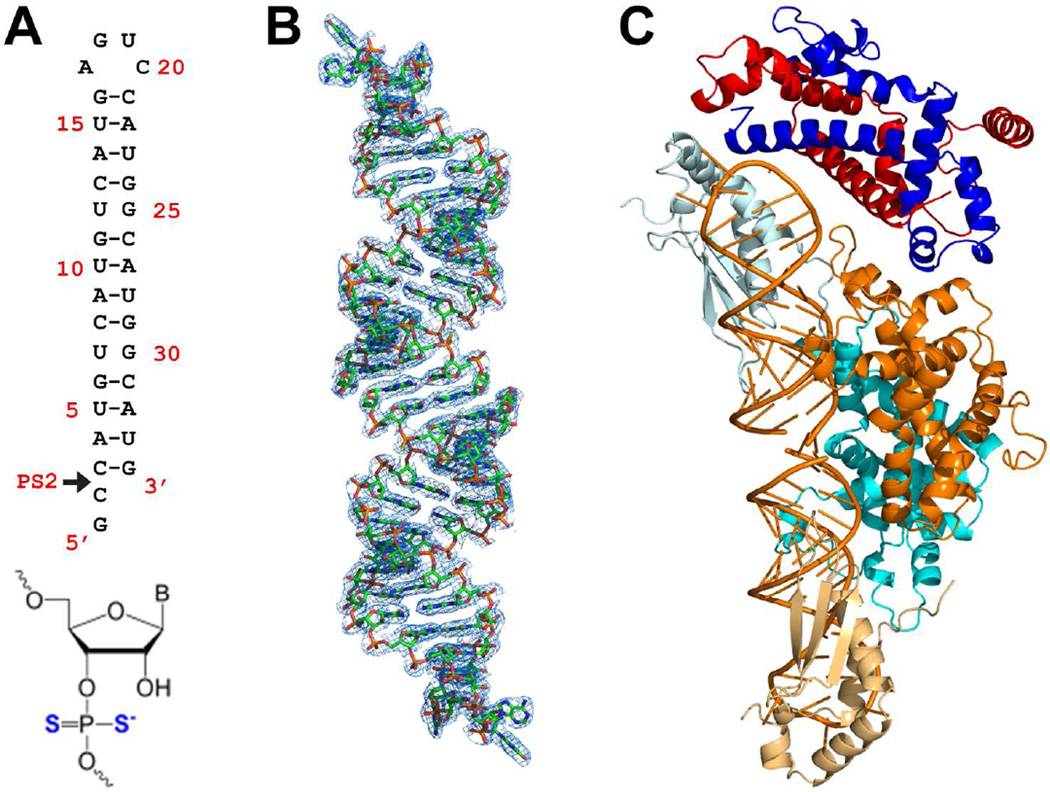
Figure 4.
Crystal structure of the Rnt1p substrate-loaded complex. (A) Sequence and secondary structure of the RNA substrate analog. The arrowhead indicates the position of the phosphorodithioated nucleotide (bottom) at the Rnt1p cleavage site. With the modification, the scissile bond cannot be cleaved by Rnt1p, and therefore, this RNA represents an Rnt1p substrate. (B) Electron density map (net in blue, 2Fo − Fc, contoured at 1σ) for the RNA pseudo-duplex (stick model in atomic colors: N, blue; C, green; O, red; P, orange; S, yellow) formed by two RNA substrate analog molecules joined with their 2-nt 5’ overhangs. (C) The architecture of the Rnt1p substrate-loaded complex. It contains one NTD dimer, one RIIID dimer, two dsRBDs, and two substrate analog RNAs in addition to solvent molecules (not shown). The protein domains are color coded as in Figure 1C and the RNA is in orange. The extra NTD dimer is not shown because it is a crystallization artifact.