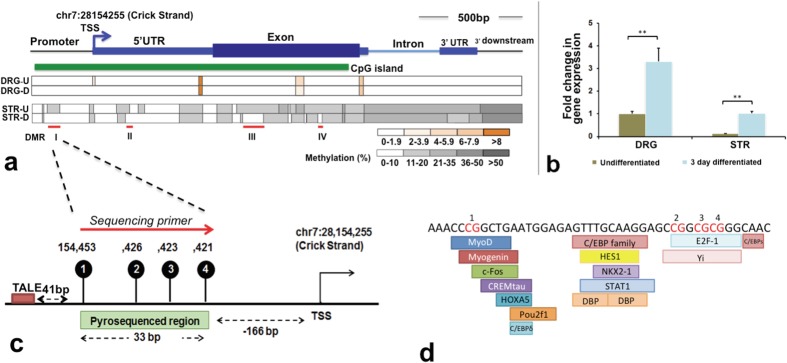
Figure 1. DNA methylation and gene expression of rat Ascl1 in DRG and STR NSCs.
(a) The gene structure of Ascl1 is shown on top of the figure. The dark blue box represents the exon, light blue boxes are untranslated regions (UTR), the blue line represents the intron, arrow indicates the direction of transcription, and the green box is CpG island. DNA methylation profiles for DRG and STR NSCs at both undifferentiated and 3-day differentiated stages (n = 3) were laid out corresponding to the gene structure. We sequenced a 2.3 kb region that spans the Ascl1 gene body and promoter regions (chr7: 28152247-28154486). The color of the box indicates the level of DNA methylation per the scale bar at the bottom of the figure. Red lines indicate statistically significant DMRs (p < 0.001) between STR-U (undifferentiated STR) and STR-D (3-day differentiated STR). (b) Relative gene expression level of Ascl1 in undifferentiated and 3-day differentiated DRG and STR neural stem cells (n = 4). The relative expression of Ascl1 was calculated using the ΔΔCt value. The average Ct values of target genes were normalized to the average Ct value of the internal control gene 18 s to calculate ΔCt, and the ΔΔCt value was generated by normalizing the ΔCt value of each cell line to the ΔCt value of the undifferentiated DRG. The p-values between any two groups, except for undifferentiated DRG and differentiated STR, were all below 0.001. (c) Pyrosequenced region in the DMR-I of Ascl1 promoter contained 4 CpG sites. The first CpG site is 41 bp upstream of the TALE binding region. The sequenced region is 166 bp from the transcription start site (TSS) of Ascl1 (d) Predicted transcription factor binding sites in the sequenced region. **P < 0.01.