Figure 4.
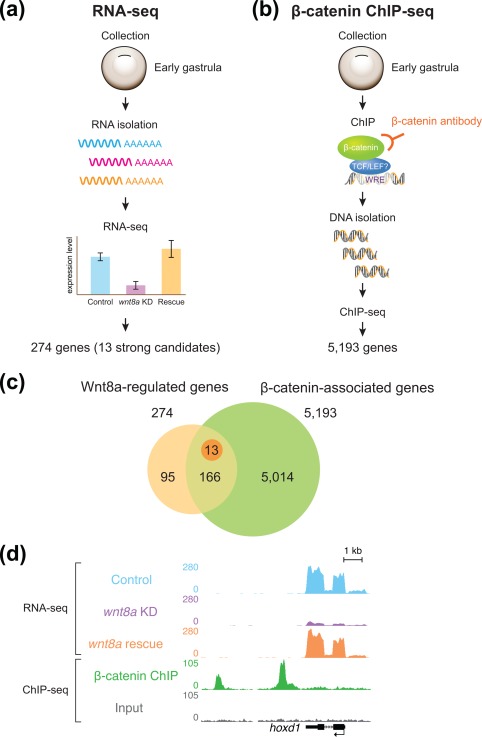
Identification of direct context‐specific Wnt/β‐catenin target genes in a genome‐wide approach. (a) Transcriptome analysis (with RNA‐seq) of early gastrula‐stage embryos. Comparing the transcriptome of control embryos with embryos with wnt8a knockdown and those with experimentally reinstated Wnt8a identifies genes regulated by Wnt8a signaling in post‐MBT embryos. (b) ChIP‐seq analysis with a β‐catenin‐specific antibody identifies DNA‐sequences associated with β‐catenin and nearby potentially regulated genes in early gastrula‐stage embryos. (c) Venn diagram comparing Wnt8a‐regulated genes (in beige) from the RNA‐seq analysis (see panel A) with the β‐catenin associated genes (in green) from the ChIP‐seq analysis (see panel B) with the overlap or intersection identifying direct Wnt8a target genes. Note that there are many more identified β‐catenin associated genes than identified Wnt8a signaling‐regulated genes in early gastrula embryos. (d) Example of a direct Wnt8a‐target gene (hoxd1) in genome view, with from top to bottom, transcripts from control embryos, transcripts from the wnt8a knock down (note reduced expression), transcripts from embryos with experimentally reinstated Wnt8a expression, β‐catenin‐associated DNA sequences (“β‐catenin” peaks), sequences of the control sample of the ChIP‐seq experiment (Input control) and the hoxd1 gene model. Note β‐catenin‐associated DNA sequences downstream (left) of the rest of the hoxd1 gene and transcript sequences of particularly the exon sequences in the control and wnt8a rescue samples
