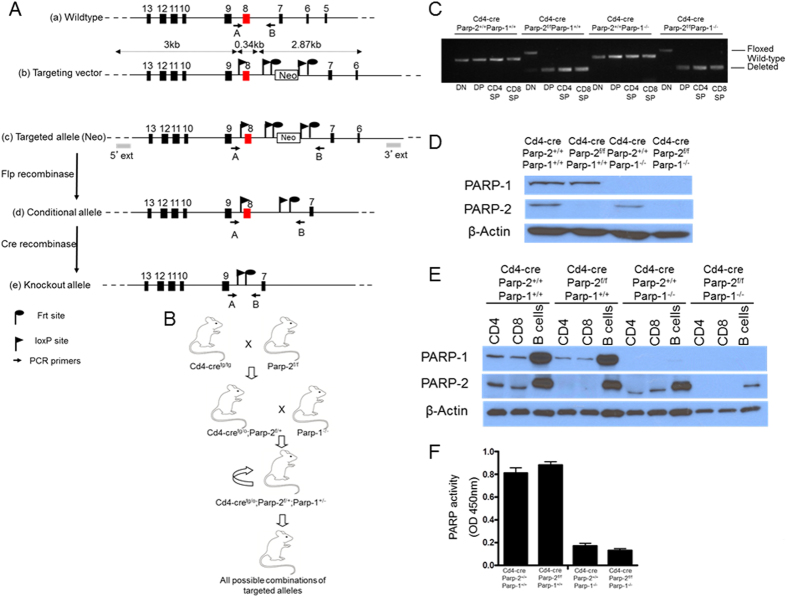
Figure 1. Generation of mice with a T-cell-specific deletion of PARP-2 in a PARP-1-deficient background.
(A) Schematic representation of (a) the wild-type allele of the mouse Parp-2 gene and the location of the genotyping primers A and B; (b, c) the structure of the correctly targeted allele with the introduced neomycin resistance cassette and loxP and FRT sites, and the locations of genotyping primers; (d) the conditional allele (flox) produced by Flp-enhanced recombinase-mediated recombination of FRT sites flanking Neo and the locations of genotyping primers; and (e) the deleted allele produced by cre recombination of loxP sites surrounding exon 8 and the locations of genotyping primers. (B) Schematic representation of the cross-breeding performed to generate mice with a T-cell-specific deletion of PARP-2 in a PARP-1-deficient background. Parp-2 floxed (Parp-2f/f) mice were crossed with Cd4-cre-transgenic mice, producing heterozygous offspring that were then crossed with Parp-1−/− mice. Cd4-cretg/o;Parp-2f/+;Parp-1+/− mice were subsequently intercrossed, producing all possible combinations of Parp-2, Parp-1 and Cd4-cre targeted alleles. (C) PCR analysis from genomic DNA in thymic double-negative (DN: CD4−CD8−), double-positive (DP: CD4+CD8+), CD4SP (CD4+CD8−), and CD8SP (CD4−CD8+) sorted subsets from Cd4-cre;Parp-2+/+;Parp-1+/+, Cd4-cre;Parp-2f/f;Parp-1+/+, Cd4-cre;Parp-2+/+;Parp-1−/−, and Cd4-cre;Parp-2f/f;Parp-1−/− mice. (D) Western-blot analysis of PARP-1 and PARP-2 protein expression in sorted DP thymocytes and (E) in sorted CD4+, CD8+, and B-cells from spleen. (F) PARP activity in protein extracts from spleen T-cells after in vitro activation with anti-CD3 + anti-CD28. Resting T-cells were isolated from spleen, cultured 14 h in the presence of anti-CD3 + anti-CD28 monoclonal antibodies, lysed and PARP activity determined in protein extracts. Results represent the mean ± SEM of a representative experiment from two independent experiments carried out in triplicate.