Figure 2.
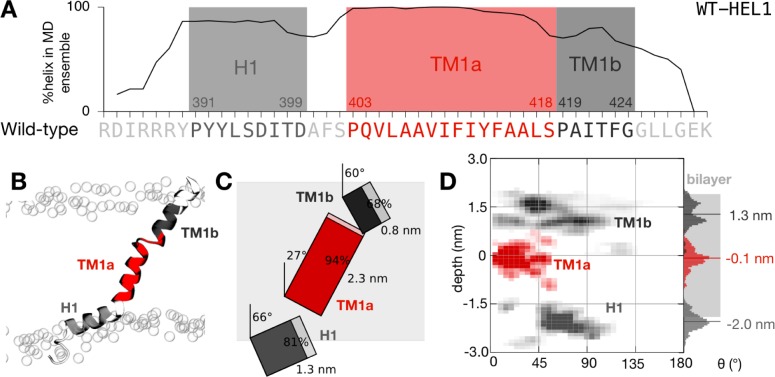
The wild-type TM1 peptide is highly dynamic and samples an ensemble of conformations. (A) The peptide remains mostly helical during the ensemble of 50 filtered simulations. As expected, the proline residues initiate regions of high helicity. We therefore defined three segments that each started from one of the three proline residues: an interfacial amphipathic helical segment (H1) and two transmembrane helical segments (TM1a and TM1b). (B) Illustrative snapshot taken from the end of the one of the molecular dynamics simulations. The peptide is colored according to panel A. The positions of the phosphate groups of the lipid bilayer are denoted with empty circles. Note the distortion of the inner leaflet of the bilayer induced by the peptide. (C) To-scale schematic representation of the average conformation with segment tilts, lengths, and helicity annotated. The average extent of the lipid bilayer as defined by the positions of the phosphate atoms is shaded gray. (D) Density plot showing the variation in the depth and tilt angle of the three segments of the peptide. The center of mass of TM1a (colored red) remains close to the center of the lipid bilayer at Phe411, and the segment explores tilt angles between 0° and 50°, centered around ∼20°.