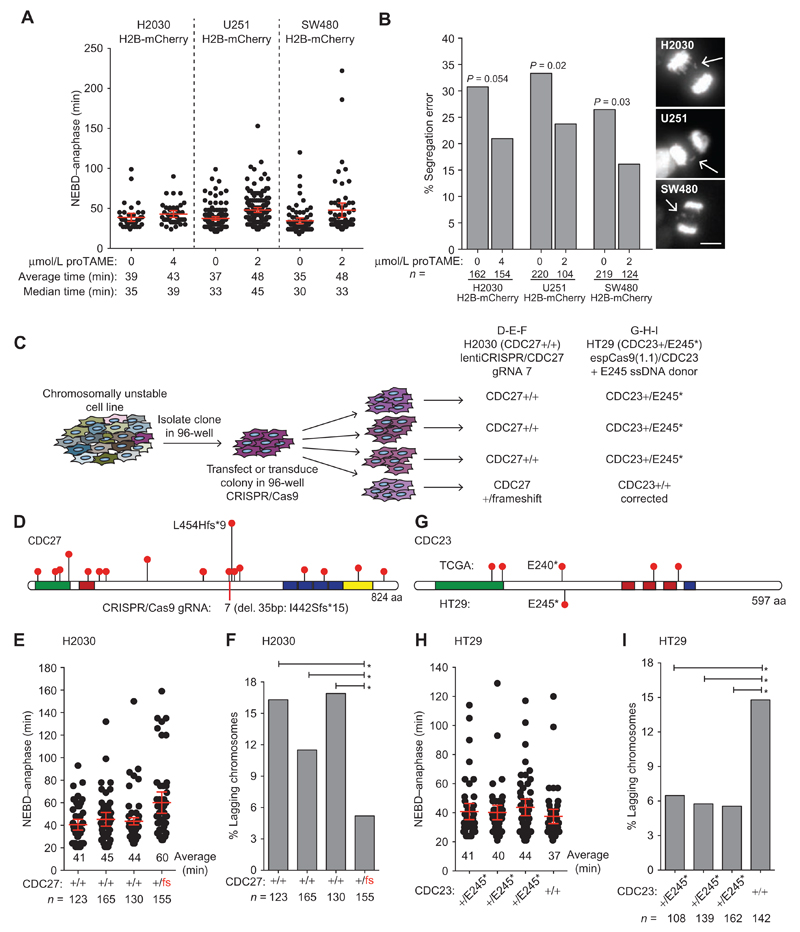
Figure 6. APC/C subunit mutational status affects CIN in cancer cells.
A and B, H2B-mCherry was introduced in H2030, U251, and SW480 cell lines and NEBD–anaphase duration (A) as well as the frequency of anaphase segregation errors (B) with and without proTAME was determined by time-lapse fluorescence microscopy. Stacks were acquired every 3 minutes, and an example of segregation error is shown for each cell line (scale bar, 10 μm; in A, bars, mean ± 95% CI of a representative experiment; in B, P values from Fisher exact test). C, Experimental procedure used to generate CRISPR/Cas9 edited H2030 and HT29 cells used for plots D to I. In each case, a single clone was infected (lentiCRISPR/CDC27 for H2030) or transfected (espCas9(1.1)/CDC23 + ssDNA donor) in a well of a 96-well plate before the colony reached confluency. Following transfection or transduction, the colony was dispersed by limiting dilution into 96-well plates. Clones were then screened for heterozygous disruption of CDC27 in H2030 cells or correction of the heterozygous E245 nonsense mutation in HT29 cells. Nonedited clones identified during screening were used as controls. Phenotypic analysis of all newly derived cell lines was performed following minimal clonal expansion to limit phenotypic diversity that may be acquired due to ongoing CIN. D, Lollipop plot of CDC27 showing only truncating mutations reported in TCGA and the location of the guide RNA used to disrupt CDC27 in H2030 cells. The clone isolated contained a heterozygous 35-bp deletion creating the truncation I442Sfs*15. E, NEBD–anaphase duration was determined for the H2030 clones using phase–contrast time-lapse microscopy (3 minutes/frame; bars, average ± 95% CI). F, The frequency of anaphase lagging chromosomes was determined on fixed cells by indirect IF microscopy (*, P < 0.05; Fisher exact test). G, Lollipop plot of CDC23 showing only truncating mutations reported in TCGA and the HT29 nonsense mutation from HT29 cells. H, NEBD–anaphase duration was determined for HT29 clones using phase–contrast timelapse microscopy (3 minutes/frame; bars, average ± 95% CI). I, The frequency of anaphase lagging chromosomes was determined on fixed cells by indirect IF microscopy (*, P < 0.05; Fisher exact test).