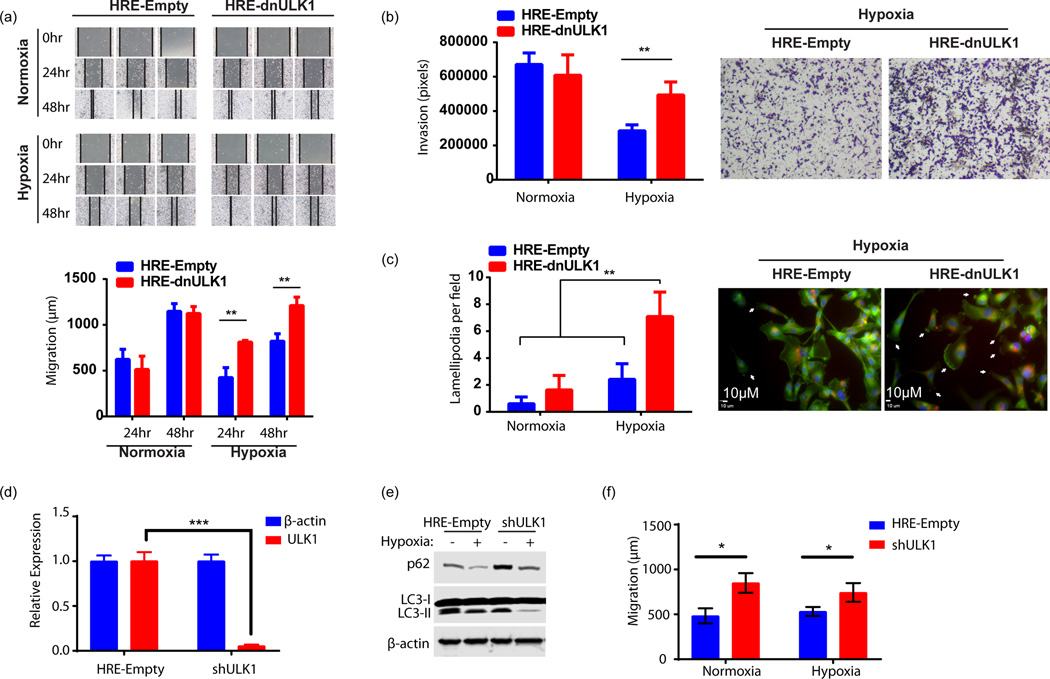
Figure 5. Loss of autophagic function in hypoxia increases invasion and migration, in vitro.
a, Wound healing assay assessing migration of MDA-MB-231 cells expressing either HRE-Empty or HRE-dnULK1 over 48hrs under hypoxic (1% O2) and normoxic conditions. Quantification was performed by measuring the distance between the two leading edges using imageJ software (mean ± s.d., n=3, **p<0.01, t-test). b, Transwell migration assay assessing invasion of MDA-MB-231 cells expressing either HRE-Empty or HRE-dnULK1 through a matrigel matrix over 48hrs under hypoxic (1% O2) and normoxic conditions. Quantification was performed using imageJ software (mean ± s.d., n=4, **p<0.01, t-test). c, Assessment of actin cytoskeletal dynamics by Alexo flour 488 conjugated phalloidin staining of MDA-MB-231 cells expressing either HRE-Empty or HRE-dnULK1 incubated in either normoxia and hypoxia (1% O2) (200× Magnification) (Blue = DAPI, Red=mCherry, Green=Alexa flour 488). Observed lamellapodia are indicated by white arrows. Lamellapodia were quantified by counting the number of cells displaying lamellapodia within a field, and averaging 15 randomly selected fields (mean ±s.d, **p<0.01, t-test). d, qPCR Assessment of ULK1 gene knockdown by shULK1 in MDA-MB-231 cells (mean ±s.d, n=3,***p<0.001, t-test). e, Immunoblot assessing autophagy markers in shRNA MDA-MB-231 cells versus HRE-Empty control. f, Wound healing assay assessing migration of MDA-MB-231 cells expressing either HRE-Empty or shULK1 over 24hrs under normoxic and hypoxic (1% O2) conditions. Quantification was performed by measuring the distance between the two leading edges using imageJ software (mean ± s.d., n=3 (normoxia) and 6 (hypoxia), *p<0.05, ns=no significance, t-test).