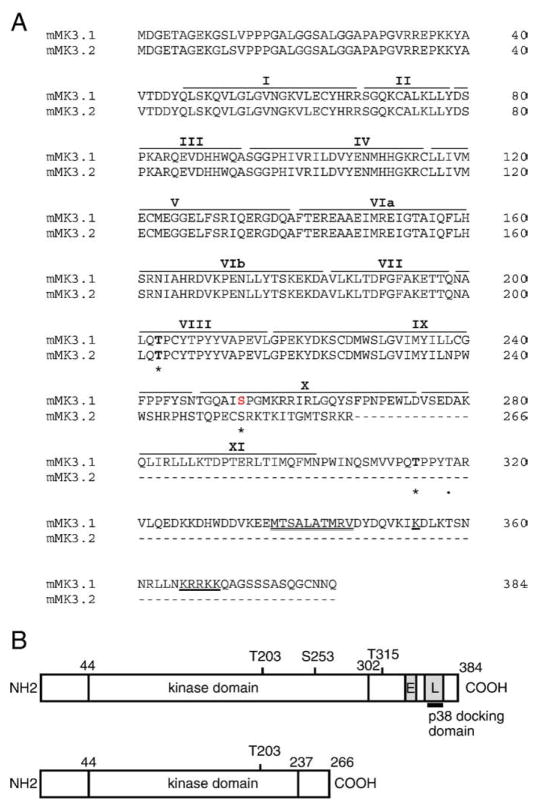
Fig. 1.
cDNA and amino acid alignment of MK3 splice variants. (A) Predicted amino acid sequence of MK3.1 and MK3.2. Putative sites of phosphorylation by p38 MAPK (Thr-203, Ser-253, and Thr-315) are indicated by an asterisk and the auto-phosphorylation site (Thr-319) is indicated by a dot. Roman numerals above refer to the subdomains (I–IX) conserved in all protein kinases. The consensus bipartite nuclear localization signal is underlined whereas the putative nuclear export sequence is indicated by a double underline. Dashes represent gaps. (B) Schematic representation of MK3.1 and MK3.2. ‘E’ nuclear export signal; ‘L’, nuclear localization signal.