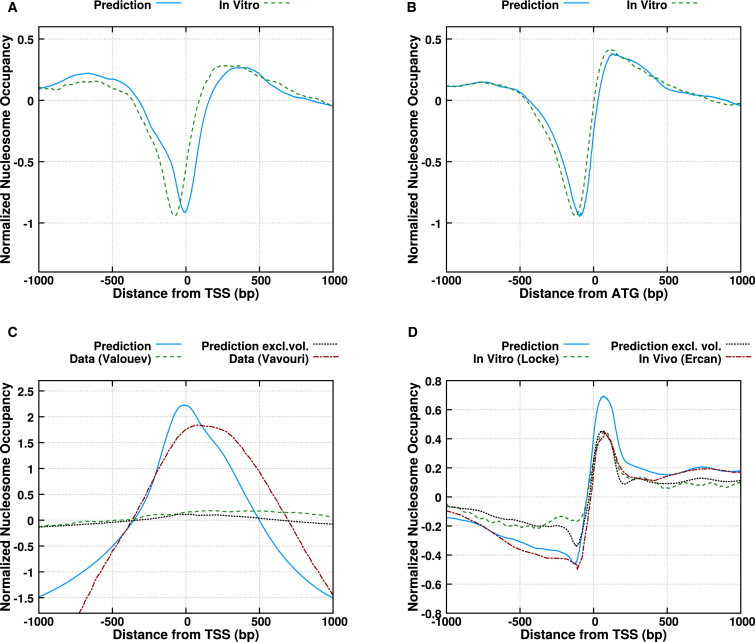
Figure 1.
Comparison of predicted and measured intrinsic nucleosome positioning signals in promoter regions. The quantities plotted are the natural logarithms of the occupancies and the signals have been normalized such that they average to zero. (Solid blue curves) Our predictions in the limit of low nucleosome density, which give an account of the strength of the signals intrinsically encoded; (dashed green curves) in vitro measurements; (dotted black curves) predictions taking into account the steric interactions. Using the same treatment as in Chevereau et al. (44), these curves have a free parameter , i.e., the difference between the chemical potential and the average energy of the landscape, which we determined to be −8.5 kT for yeast (curves not shown due to similarity with the low-density limit), −5.7 kT for C. elegans, and −1.38 kT for humans. (A and B) S. cerevisiae, average nucleosome occupancy centered on the TSS and start codons, respectively. Data from Kaplan et al. (18). (C) Like (A), for Homo sapiens. The in vitro data is from Valouev et al. (34). Additionally shown is the nucleosome retention signal from Vavouri and Lehner (36). (D) Like (A), for C. elegans. The in vivo data is from Ercan et al. (24); the in vitro data is from Locke et al. (38). To see this figure in color, go online.