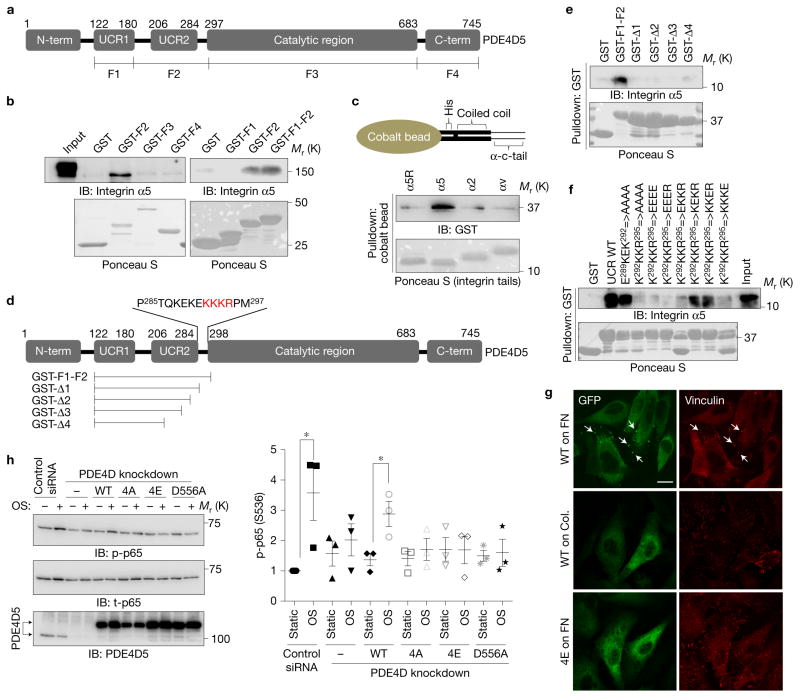
Figure 5.
Mapping the integrin binding site on PDE4D5. (a) Schematic representation of PDE4D5 and fragments used for pulldown assays. (b) HUVEC lysates were incubated with GST-tagged fragments of PDE4D5 and probed for integrin α5. Results are representative of three independent experiments. (c) To test whether the interaction is direct, integrin α tails immobilized on cobalt beads were incubated with purified PDE4D5 F2 fragment. Beads were washed and bound material was analysed by western blotting (α5R: scrambled sequence of α5 tail). (d) Deletion constructs used for detailed mapping and critical residues for binding. (e,f) The indicated PDE4D5 fragments and mutants were immobilized on GSH beads and incubated with the α5 tail protein used in c. Bound α5 tail protein was detected by western blotting with integrin α5 antibody against cytoplasmic tail. Results are representative of three independent experiments. (g) BAECs expressing GFP-tagged PDE4D5 WT or the 4E mutant were plated on FN or collagen and sheared for 15 min. The cells were fixed and stained for the focal adhesion marker, vinculin. Arrow indicates colocalization of PDE4D5 with vinculin. Results are representative of three independent experiments. Scale bar, 50 μm. (h) BAECs stably expressing integrin α5 binding-deficient PDE4D5 mutants (4A and 4E) or the catalytically inactive mutant (D556A) were transfected with siRNA to knock down the endogenous PDE4D, and then were subjected to oscillatory shear for 18 h. NF-κB activity was assayed as in Fig. 1; (n= 3 independent experiments). Data are represented as means ± s.e.m. *P < 0.05 by two-tailed t-test. Source data are provided in Supplementary Table 1. Unprocessed scans of blots are shown in Supplementary Fig. 6.