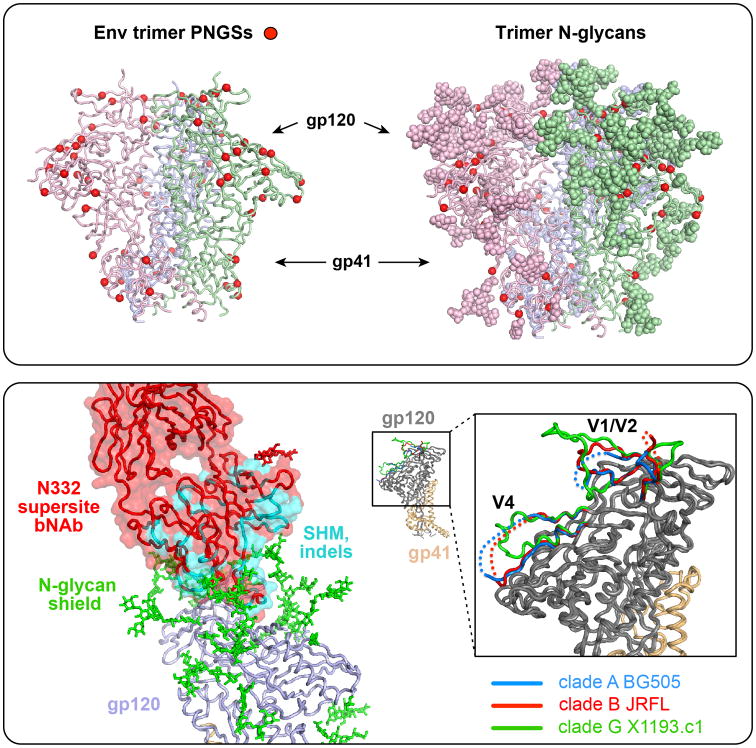
Fig 1. N-glycan shielding, trimer packing and variability.
Top box, left; Potential N- glycosylation sites (PNGS; motif is NXT/S) are shown (red balls) on the structure of the HIV trimer spike (BG505 SOSIP.664). Each of the three protomers, composed of gp120 and gp41 associated subunits are shown with matching colors for each monomer (hot pink, blue and green, left, back and right, respectively). Note the “spring-loaded” helices of gp41 can be seen in the center of the lower region of the trimer (and in the lower box, lower right inset). Right, the N-glycans are shown in a space-filling, ball rendition, “shielding” the underlying protein polypeptide surface; asparagine residues at the N-glycan base are indicated (red balls). Lower box, lower left, the bNAb PGT121 (red, surface and alpha carbon backbone) penetrates the N-glycan shield (green) with the gp120 alpha carbon trace in blue (ribbon). Somatic hypermutation and indels are indicated (cyan), achieving a relatively high level of 23% in this bNAb, many of which are needed to allow mAb penetration of the evolved dense N-glycan shield that accounts for half the molecular mass of the HIV Env trimer. Lower right, smaller image is one gp120-gp41 protomer with gp120 (gray) associated with the spring-loaded gp41 (gold); expanded box depicts the variable regions V1/V2 and V4 of gp120 from three strains derived from different subtypes as indicated. These V regions, along with N-glycan shielding, contribute to immune evasion from neutralizing antibodies by presenting variable surfaces that can accommodate additional mutation.