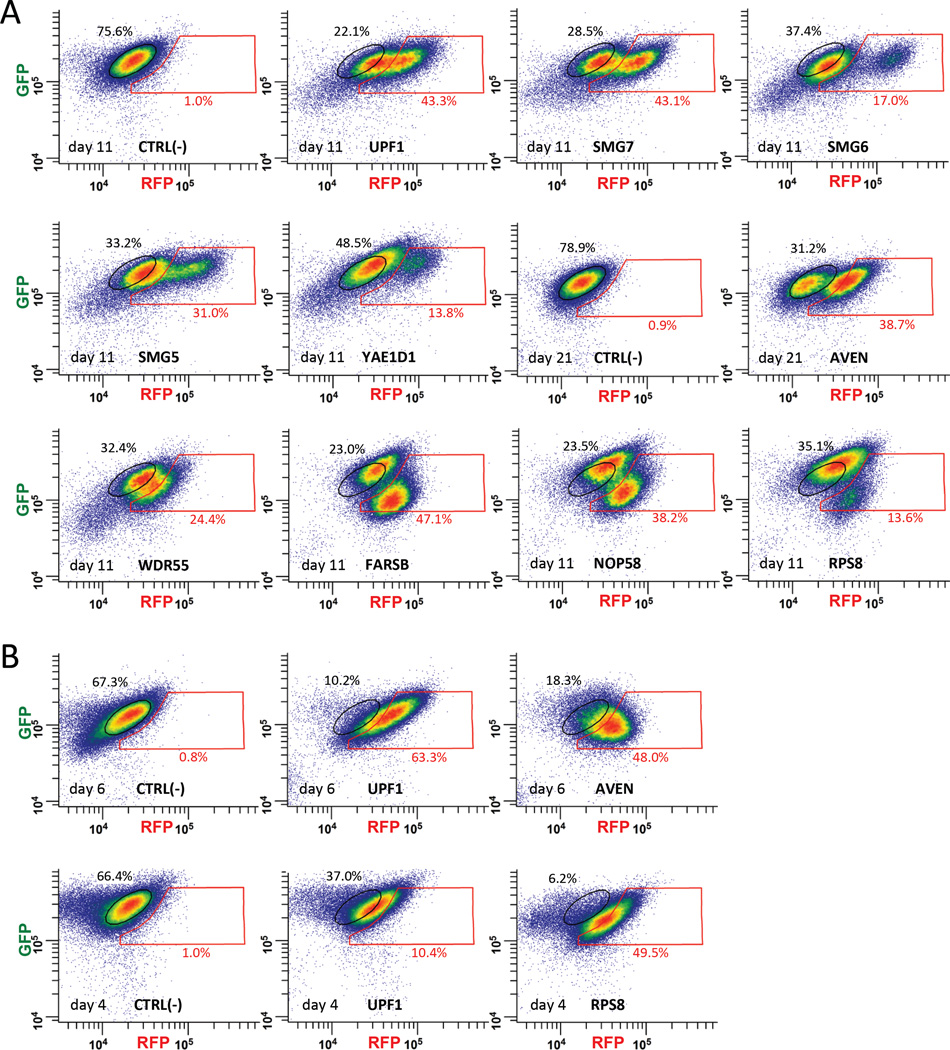
Figure 3. FACS analysis of the fluorescence shift produced by individual sgRNAs identified by Fireworks screening of the GeCKO-LtCRISPR sgRNA library for factors affecting human NMD.
(A) sgRNAs obtained from the genome-wide screen (Table 1) were individually transduced into the “green” Fireworks cell line (Figure 1A) and the resulting cells were FACS-analyzed for an increase in red fluorescence. Populations of cells with increased red fluorescence (nearly not observable [0.9–1.0%] in the corresponding negative controls) appear in the red gate. Fractions (percent) of cells in the original (black gates) and shifted (red gates) populations are indicated. (B) To exclude possible off-target effects of sgRNAs, two of the genes not previously implicated in NMD, AVEN and RPS8, were targeted by shRNAs. FACS-analyses of the shRNA-transduced “green” Fireworks cell line are shown; populations of cells with increased red fluorescence appear in the red gate (FACS-analysis of the orthogonal “red” Fireworks cell line transduced with shRNA targeting RPS8 is shown in Figure S2B). See also Figure S2.