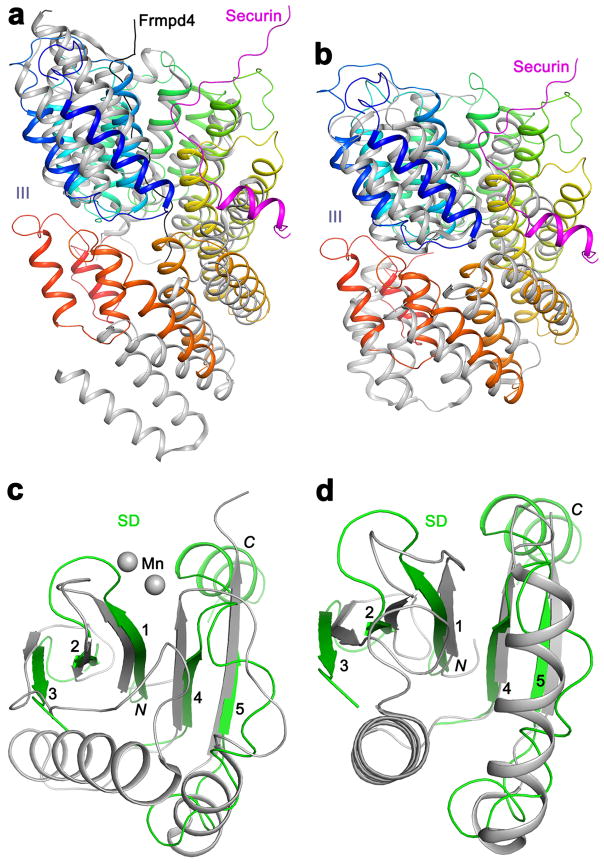
Extended Data Fig. 7.
Structural homologs of domains III and SD of separase. (a). Overlay of the structures of domain III of separase (color ramp from N- (blue) to C-terminus (red)) and the TPR domain of LGN (gray, PDB entry 4WNG; 11% sequence identity, Z score of 14.4) 23–25. The Frmpd4 ligand (black) of LGN is bound to a different region of the structure compared to securin. (b). Overlay of the structures of domain III of separase and the subunit 7 of the APC/C (PDB entry 5G04; 5% identity, 14.3 Z score) 37. (c). Overlay of the structures of the SD of separase (green) and a part of the PIWI domain of Argonaute (gray, PDB entry 4N76; 10% sequence identity, 5.5 Z score. As a comparison, matching this β-sheet to that in C. thermophilum separase produced a Z score of 5.7.) 27,38. Residues in the helical insert between β3 and β4 of separase are removed for clarity. (d). Overlay of the structures of the SD of separase and the YqgF domain of Tex (gray, PDB entry 3BZK; 4% sequence identity, 5.5 Z score) 39.