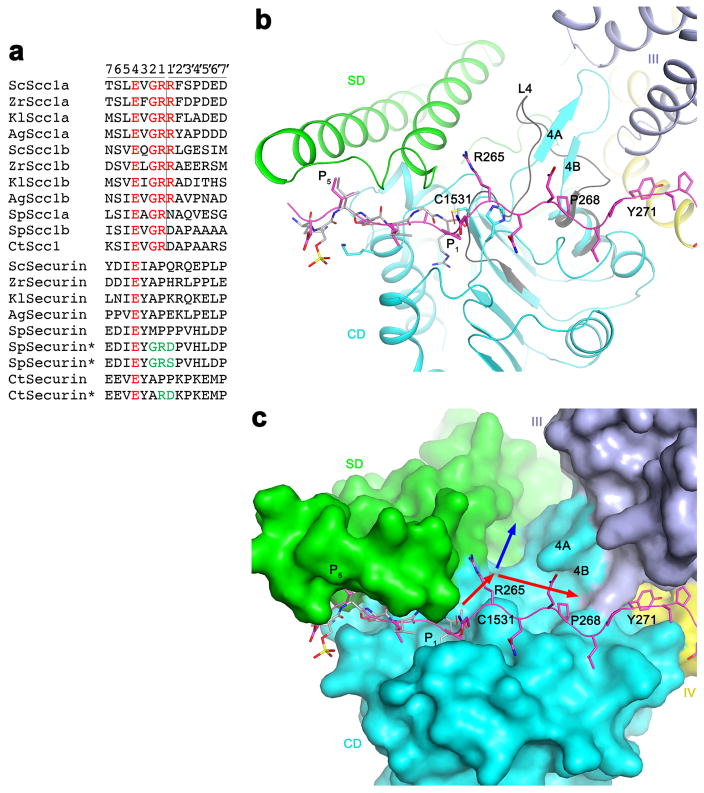
Extended Data Fig. 8.
Possible binding groove for the P′ residues of the substrate. (a). Alignment of the separase cleavage sites in Scc1 substrates. The two cleavage sites in each protein are named a and b. The equivalent residues in securin are also shown. The asterisks indicate securin mutants (mutations in green) that become substrates of separase. The P and P′ residues are labeled at the top, and the cleavage site is indicated with the vertical line. (b). The overall binding mode of residues 258–271 of securin in separase. The β4A-β4B segment of CD (cyan) is a loop (L4, dark gray) in the C. thermophilum SD-CD structure. (c). A groove in the active site of separase (red arrows) can accommodate the P′ residues. The blue arrow indicates another groove in this region, but the binding mode of securin suggests that the groove indicated by the red arrows is more likely.