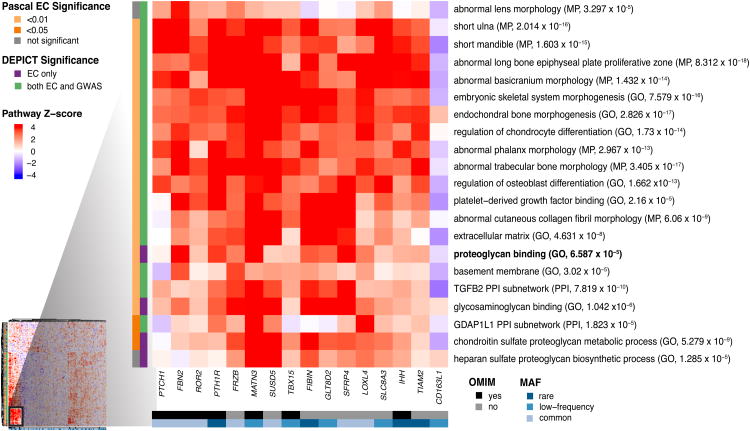
Figure 2.
Heat map showing subset of DEPICT gene set enrichment results. The full heat map is available as Extended Data Fig. 7. For any given square, the color indicates how strongly the corresponding gene (shown on the x-axis) is predicted to belong to the reconstituted gene set (y-axis). This value is based on the gene's Z-score for gene set inclusion in DEPICT's reconstituted gene sets, where red indicates a higher Z-score and blue indicates a lower one. The proteogly can binding pathway (bold) was uniquely implicated by coding variants by DEPICT and PASCAL. To visually reduce redundancy and increase clarity, we chose one representative “meta-gene set” for each group of highly correlated gene sets based on affinity propagation clustering (Supplementary Information). Heat map intensity and DEPICT P-values correspond to the most significantly enriched gene set within the meta-gene set; meta-gene sets are listed with their database source. Annotations for the genes indicate whether the gene has OMIM annotation as underlying a disorder of skeletal growth (black and grey) and the minor allele frequency of the significant ExomeChip (EC) variant (shades of blue; if multiple variants, the lowest-frequency variant was kept). Annotations for the gene sets indicate if the gene set was also found significant for EC by PASCAL (yellow and grey) and if the gene set was found significant by DEPICT for EC only or for both EC and GWAS (purple and green). Abbreviations: GO: Gene Ontology; MP: mouse phenotype in the Mouse Genetics Initiative; PPI: protein-protein interaction in the In Web database.