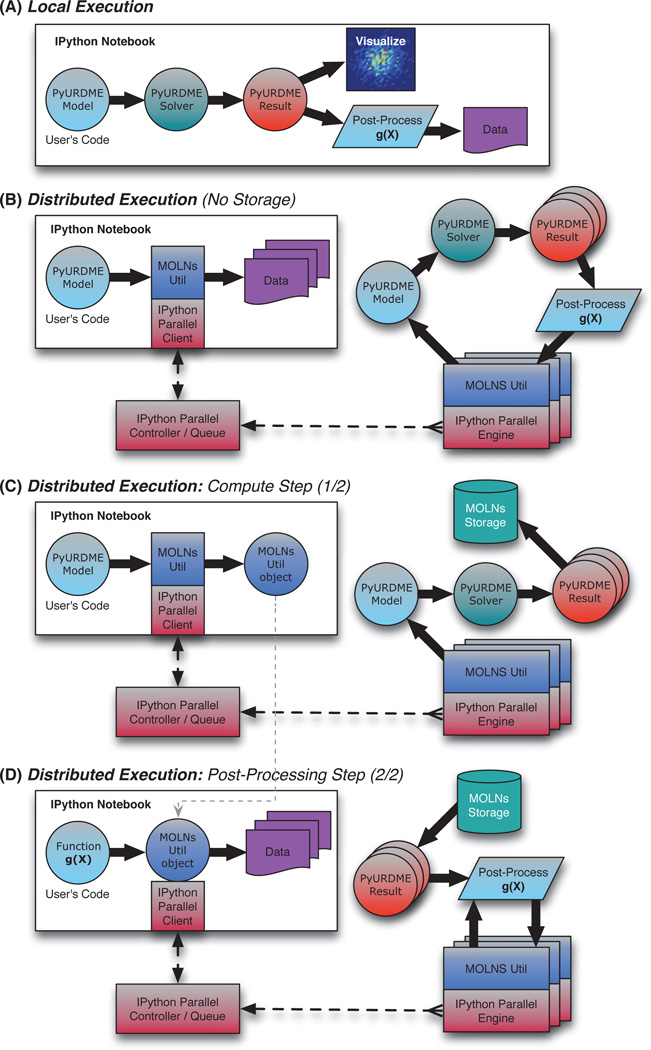
Fig. 4.
MOLNs workflows. (A) Basic workflow executed within the IPython notebook. The user develops a biological model, and the model is executed by the solver to produce a result object. The results are either visualized using functionality in PyURDME or passed to a user-defined postprocessing function g(x). This local simulation workflow does not require molnsutil and can hence be developed locally without cloud resources. (B) Distributed computational workflow. The user develops a biological model and a postprocessing function and passes them to molnsutil, which arranges the distributed execution into tasks and enacts it using IPython parallel. Each task executes the model to produce one or more result objects which are processed by the user-supplied g(x). The resulting data is aggregated and returned to the user’s IPython notebook session. (C) In many cases it is advantageous to separate the generation of the result objects from the postprocessing. This shows the distributed workflow of generating the results and storing them in the integrated storage services so that subsequent runs of the postprocessing analysis scripts (D) can be done multiple times, allowing interactive development and refinement of these scripts.