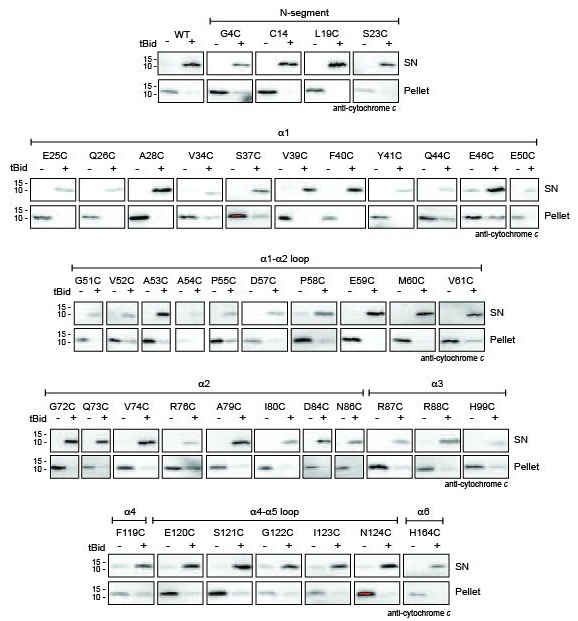
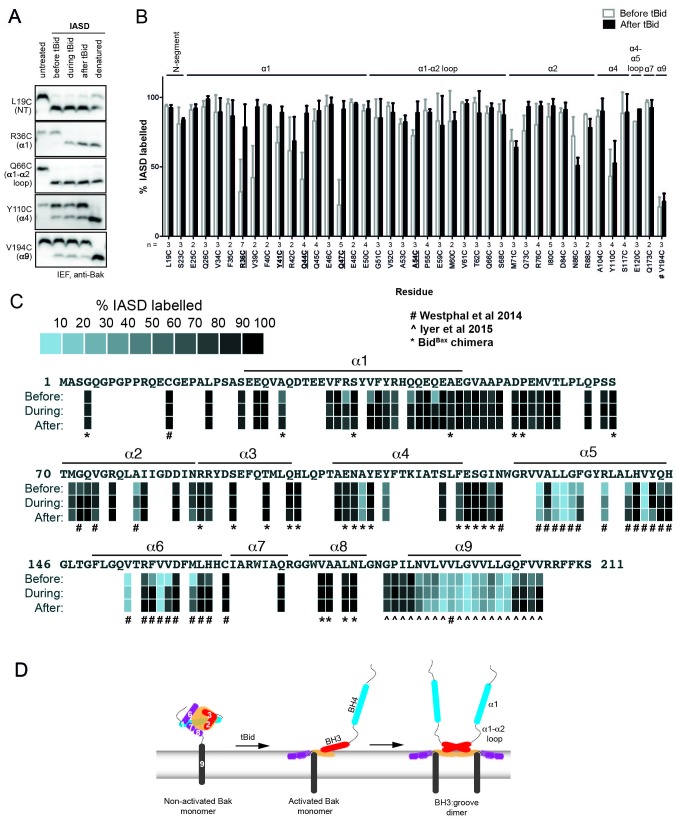
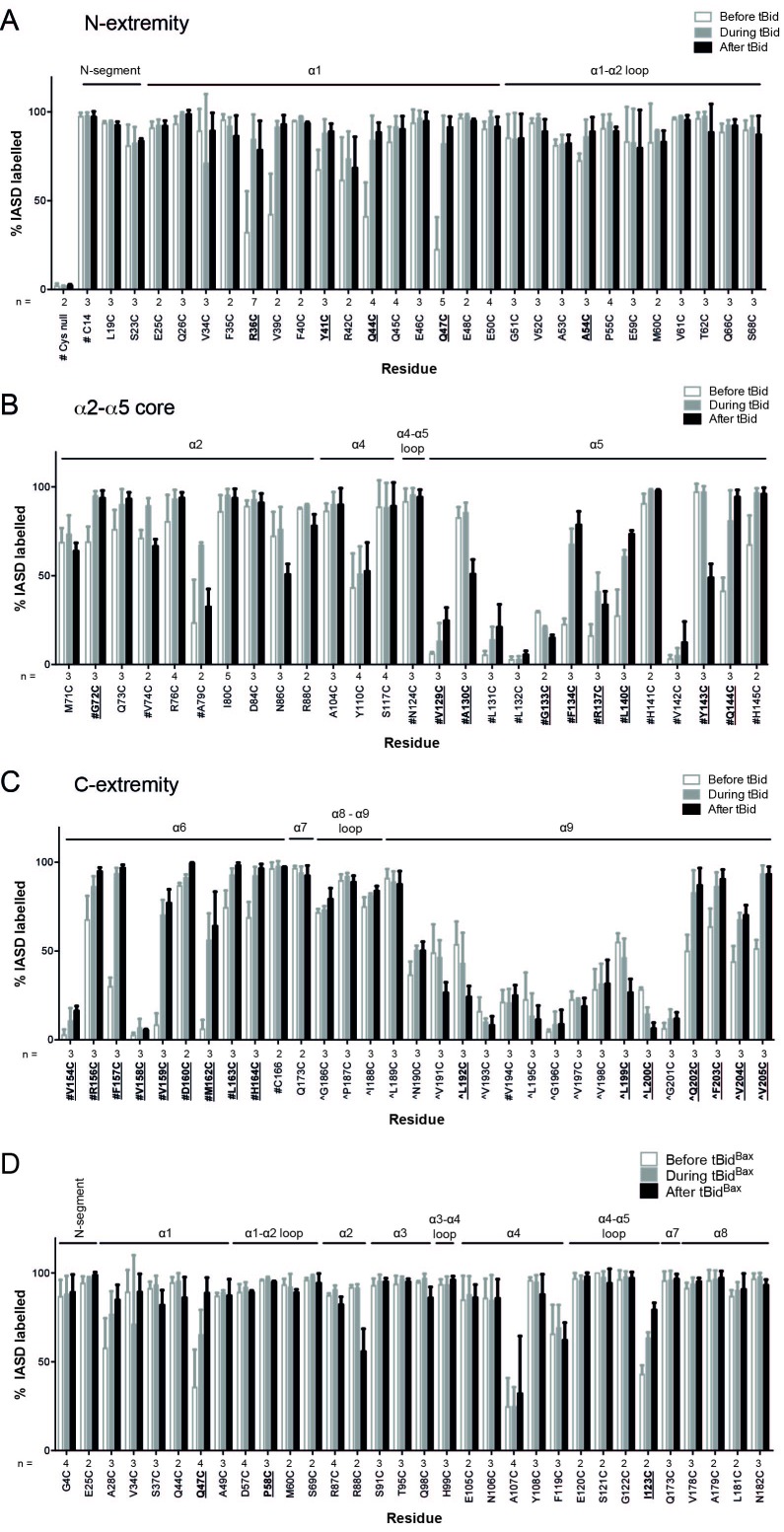
Figure 1. Following oligomerisation, the Bak N-segment, α1 and α1-α2 loop become fully solvent-exposed in contrast to the partially exposed core (α2-α5) and latch (α6-α9).
(A) Solvent exposure of hBak cysteine mutants was assessed by IASD labelling before (lane 2), during (lane 3) and after (lane 4) treatment with tBid. Controls of unlabelled (untreated, lane 1) and fully labelled (denatured, lane 5) Bak were included for comparison. Example IEF western blots are shown. (B) Quantitation of IASD labelling before and after treatment with tBid for the panel of previously untested Bak residues. Data are mean ± SD (n ≥ 3), or range (n = 2), with n for each residue labelled on the x-axis. IASD labelling data for residue V194C were from (Westphal et al., 2014) (denoted #). Residues for which there is a significant difference in IASD labelling before versus after tBid are in bold and underlined (p<0.05). (C) Heat map overview of Bak IASD labelling with tBid treatment from (B) pooled with published analyses from (Westphal et al., 2014)(denoted #) or (Iyer et al., 2015)(denoted ^ ) and from treatment with the tBidBax chimera (denoted *; see also Figure 1—figure supplement 3; Figure 1—source data 1). (D) Schematic of Bak structural rearrangement from its non-activated monomeric state to the activated dimer. Helices are numbered for the non-activated Bak. Note the complete solvent-exposure of α1 and the α1-α2 loop in the activated dimer. The following figure supplements are available for Figure 1.
DOI: http://dx.doi.org/10.7554/eLife.19944.003
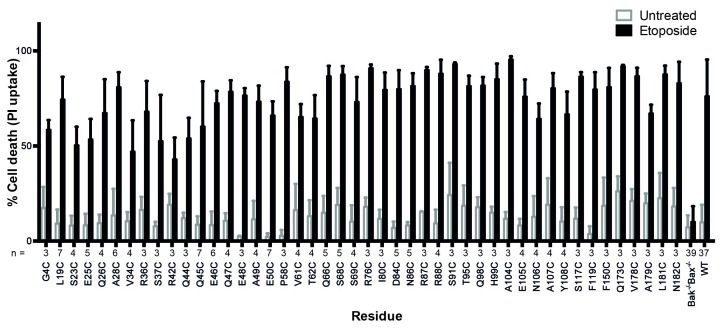
Figure 1—figure supplement 1. Bak cysteine variants retain apoptotic function.
Figure 1—figure supplement 2. Bak cysteine variants retain apoptotic function in response to tBid.