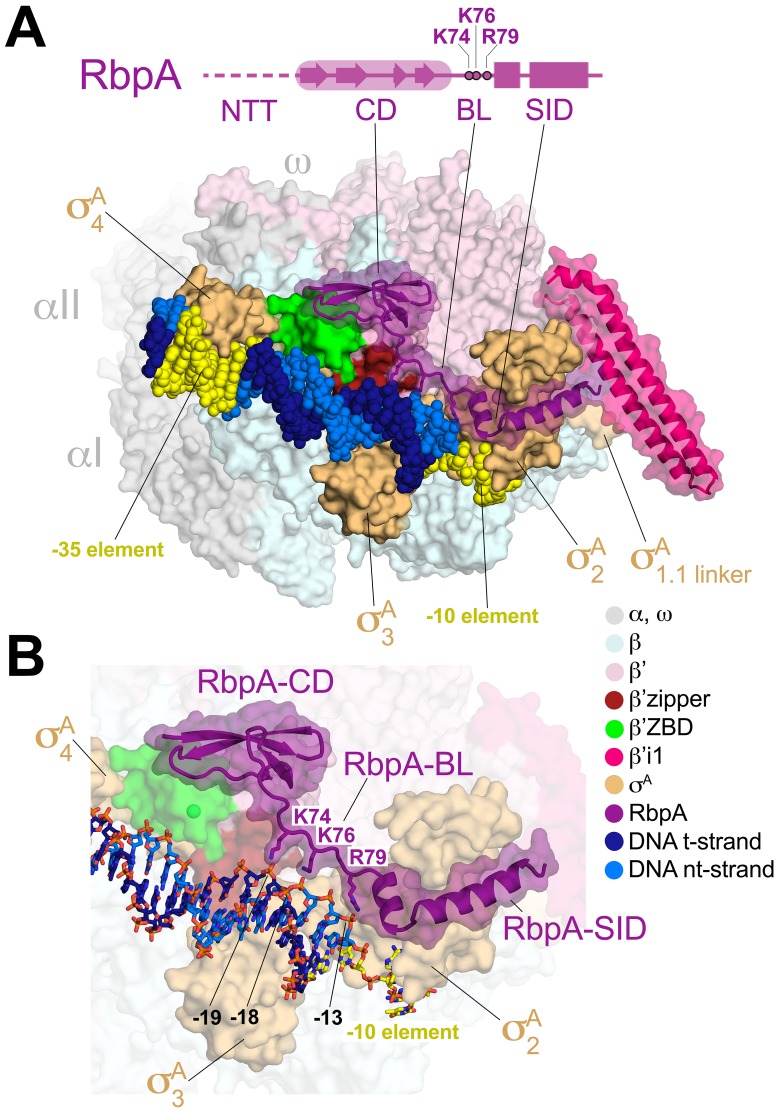
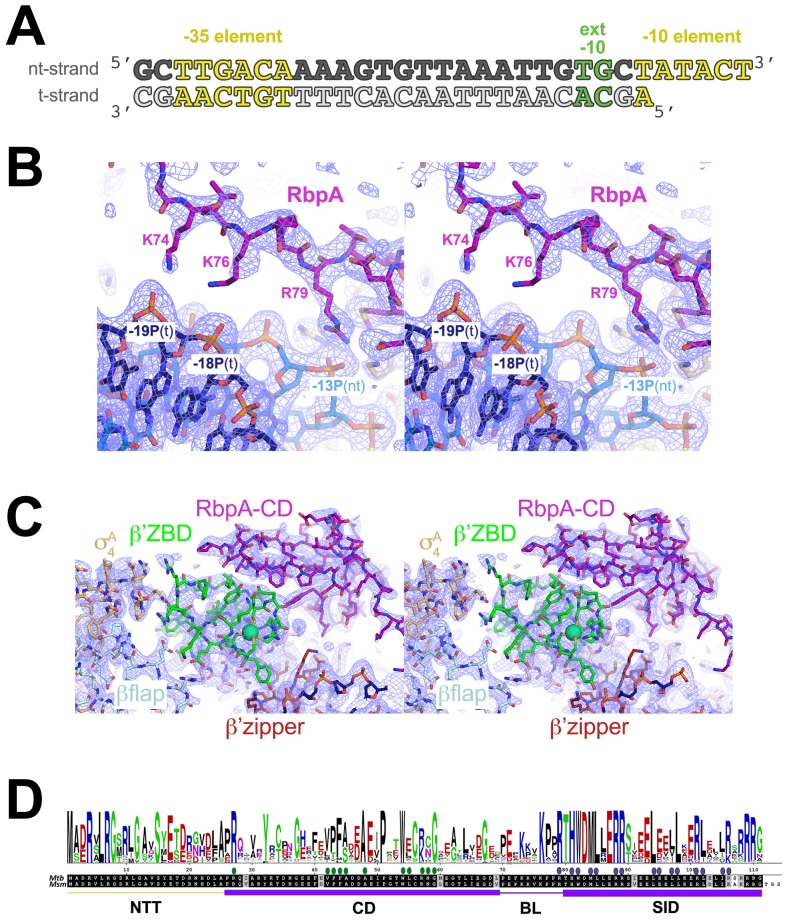
Figure 1. Structure of the Msm RbpA/TIC.
(A) (top) The RbpA structural architecture is represented schematically. The CD is shown as a thick region, with β-strands represented as arrows. The α-helices of the SID are shown as rectangles. Linker regions lacking secondary structure, the NTT and BL, are represented by a thin line. The NTT is disordered in the crystal structure and is shown as a dashed line. Conserved basic residues in the BL (K74, K76, R79) that interact with the DNA phosphate backbone are denoted. (bottom) Overall structure of the Msm RbpA/TIC. The color-coding of most of the structural features is denoted in the legend. Protein components (core RNAP, σA, RbpA) are shown as molecular surfaces. The surfaces of RbpA and the lineage-specific insert β’i1 are transparent, revealing the α-carbon backbone ribbon underneath. RbpA side chains K74, K76, and R79 are shown in stick format. The DNA is shown as CPK atoms, with the −35 and −10 elements colored yellow. (B) Magnified view of the region including RbpA and the promoter DNA near the −10 element. The DNA is shown in stick format. The β’ZBD surface is transparent with the Zn2+-ion shown as a sphere.