Figure 3. Graphical representation of clonal evolution from primary diagnosis to relapse based on targeted deep sequencing of driver mutations.
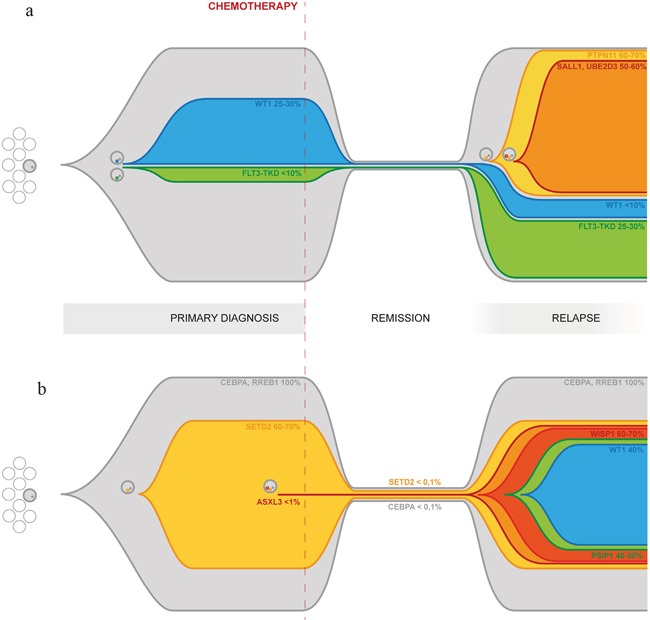
Panel 3a: Clonal evolution in patient AML#3. The primary tumor differentiates into subclones through the acquisition of new somatic mutations, including WT1 and FLT3-TKD. Those clones survive chemotherapy and contribute to relapse. Later acquisition of additional mutations, such as PTPN11, SALL1 and UBE2D3, further increases clonal heterogeneity and confers a higher degree of complexity to the disease. Reported percentages refer to the estimated size population of each clone inferred from the MF calculated on targeted deep sequencing data for each mutation and adjusted for CN. Panel 3b: Clonal evolution in patient AML#2. The entire tumor population both in the primary and in the relapse samples carries a biCEBPα and a RREB1 mutation. The inactivation of SETD2 in a substantial fraction of the cells is associated with the acquisition of a mutator phenotype causing differentiation into multiple minor subclones through the acquisition of additional somatic mutations, increasing the plasticity and adaptability of the leukemia. High coverage targeted deep sequencing was able to detect persistence of biCEBPAα and SETD2 mutation during remission. Reported percentages refer to the estimated size population of each clone inferred from the MF calculated on targeted deep sequencing data for each mutation and adjusted for CN.
