Abstract
Triatoma infestans is an important hematophagous vector of Chagas disease, a neglected chronic illness affecting approximately 6 million people in Latin America. Hematophagous insects possess several molecules in their saliva that counteract host defensive responses. Calreticulin (CRT), a multifunctional protein secreted in saliva, contributes to the feeding process in some insects. Human CRT (HuCRT) and Trypanosoma cruzi CRT (TcCRT) inhibit the classical pathway of complement activation, mainly by interacting through their central S domain with complement component C1. In previous studies, we have detected CRT in salivary gland extracts from T. infestans. We have called this molecule TiCRT. Given that the S domain is responsible for C1 binding, we have tested its role in the classical pathway of complement activation in vertebrate blood. We have cloned and characterized the complete nucleotide sequence of CRT from T. infestans, and expressed its S domain. As expected, this S domain binds to human C1 and, as a consequence, it inhibits the classical pathway of complement, at its earliest stage of activation, namely the generation of C4b. Possibly, the presence of TiCRT in the salivary gland represents an evolutionary adaptation in hematophagous insects to control a potential activation of complement proteins, present in the massive blood meal that they ingest, with deleterious consequences at least on the anterior digestive tract of these insects.
Introduction
Hematophagous insects, such as “kissing bugs,” the vectors of Chagas disease, ingest about ten times their body weight in fresh blood. Blood from mammals harbors a remarkable number of molecular and cellular strategies to destroy foreign membranes. The complement system, mainly present in blood, is a central arm of innate and adaptive immune responses. It has several macromolecules specialized in swiftly recognizing a variety of molecular nonself-patterns with subsequent activation of an array of highly efficient destructive mechanisms. It is conceivable that the hematophagous insect saliva contains molecules that counteract the potential deleterious effects that blood complement may have at least on the anterior digestive tissues in these insects. Herein, we propose that salivary TiCRT may provide the insect with such defensive mechanism.
Arthropod vectors of Chagas disease in Chile are Triatoma infestans, Mepraia spinolai, Mepraia gajardoi,1 and Mepraia parapatrica.2 Triatoma infestans is responsible for the domestic cycle of the disease, whereas M. spinolai, M. gajardoi, and M. parapatrica are responsible for the wild cycle. Chagas disease is a zoonosis that affects about 6 million people in Latin America3 and there is a growing concern because it is emerging in other continents. Thus, only in the United States, 300,000 to 1 million people are now infected.4 Nonvectorial mechanisms, such as congenital, blood transfusions, organ transplantation from donors infected with Trypanosoma cruzi, as well as ingestion of contaminated foods, are considered to be largely responsible.5
Although during the last two decades great strides in the study of T. cruzi genetics and its relationship with the host immune system have occurred, there is still no effective safe cure for this disease.
Saliva from hematophagous insects possesses different molecules that mediate a successful feeding process. They counteract the hemostatic, inflammatory, and immune processes of the host,6 such as vasoconstriction, blood coagulation, and platelet aggregation,7–11 preventing the blood loss that follows tissue injury. However, the success of the hematophagic process also depends on innate and adaptive immune responses and, specially, on the complement system.12
The vertebrate complement system is a component of the immune response that has approximately 30 soluble and integral membrane proteins.13 It has three main macromolecular recognition modules (C1, mannan-binding lectin [MBL], and ficolins) that recognize diverse microorganism-associated molecular patterns (“danger signals”), through three activation routes. The classical pathway can be initiated by the binding of C1 directly to the pathogen surface or during an adaptive immune response, by the binding of C1 to antibody–antigen complexes, being a key link between effector mechanisms of innate and adaptive immunity. The MBL pathway is initiated by the binding of MBL, a serum C1-like molecule, to mannose-containing carbohydrates on bacteria or viruses. The ficolin pathway is initiated by the binding of ficolins, also serum C1-like molecules, to a variety of lipoteichoic acid–like structures on the membrane of aggressor cells. Finally, the alternative pathway is initiated when spontaneously low-rate activated C3b-like molecules bind to permissive membranes. The presence of sialic acid on host cells allows the operation of regulatory molecules that will inactivate these molecules. Since most microbial membranes are devoid of sialic acid, the C3b-like molecules remain capable of recruiting factor B, thus constituting a functional C3-convertase. Derived from these three activation pathways, a cascade of pro-inflammatory, opsonizing, biological membrane–destructive and costimulating of immune response functions, among others, is unleashed.14
Calreticulin (CRT) is a protein that has many functions in mammals.15,16 Structurally, it is divided in three domains: An N-terminal, the most conserved among CRTs15; a P-internal, binds calcium with high affinity; and a C-terminal, highly acidic, binds calcium with high capacity.17 Of utmost importance in the studies described here, an S domain, located between N and P, binds C1, and neutralizes its capacity to activate C4, an essential step necessary for the initiation of the classical pathway activation.18 Human and T. cruzi CRTs specifically bind C1, inactivating at least two of the three described pathways.19 On the other hand, salivary gland extracts from hematophagous insects such as Lutzomyia longipalpis, Lutzomyia migonei, Panstrongylus megistus, Triatoma brasiliensis, and Rhodnius prolixus inhibit the complement system, thus favoring the feeding process.20 Moreover, it has been probed that T. brasiliensis forced to ingest serum in conditions in which the protection of midgut by salivary inhibitors is bypassed, and showed damage by complement in the anterior midgut epithelium and cell death.12
To address the role of salivary CRT on the hematophagous feeding process, we have cloned and characterized TiCRT and its S domain (TiS). Herein, we show that recombinant TiS (rTiS) binds human C1 and inhibits the classical pathway of complement activation in vitro. Conceivably, CRT in insect saliva protects the anterior digestive tract of these insects against deleterious effects of the complement system.
Methods
Experimental insects.
Adult live T. infestans (Hemiptera: Reduviidae) were reproduced in one of our laboratories (Werner Apt). The insects were maintained at 28°C, 65% relative humidity in a dark room. They were fed weekly on chickens (Gallus gallus). The insects were anesthetized and the soft parts were obtained by dissection under 10× magnification and kept on RNA Later (Qiagen, Germantown, MD).
Definition of the functional state of the complement system in blood ingested from T. infestans.
Blood from T. infestans was recovered by introducing a syringe needle into its anterior midgut, immediately after feeding on a chicken. As this blood does not coagulate, plasma was obtained. Microtitration plates (Nunc MaxiSorp; Sigma-Aldrich, St. Louis, MO) were coated with 100 μL of human IgM, at 4 μg/mL (Jackson ImmunoResearch, West Grove, PA) and incubated at 4°C overnight. Nonspecific binding sites were blocked with 250 μL of 3% w/v bovine serum albumin in phosphate-buffered saline (PBS). Then, the following was added: 1) 100 μL of chicken normal serum (CNS) at a 1:320 dilution, as a source of active complement C1. CNS was obtained from the same bird that was used to feed the insects; 2) 100 μL of blood obtained from the insect anterior midgut at a 1:320 dilution; 3) 100 μL of CNS at a 1:320 dilution, plus ethylenediaminetetraacetic acid (EDTA) 10 mM final concentration; and 4) 100 μL of undiluted blood from the insect's anterior midgut plus EDTA 10 mM (3 and 4 were positive inhibition controls). They were incubated for 1 hour at 37°C with human C4 in Veronal Buffer (5,5-diethylpyrimidine-2,4,6[1H,3H,5H]-trione) at pH 7.3. C4b deposits, an indication of complement activation, were detected with an affinity-purified rabbit anti-human C4 antibody (DAKO, CA) at a 1:4,000 dilution, followed by an affinity-purified horseradish peroxidase (HRP)–conjugated goat anti-rabbit IgG antibody (DAKO, Carpinteria, CA). HRP activity (measured at 405 nm) was assessed by addition of 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulphonic acid) diammonium salt (ABTS) with 0.03% v/v H2O2. The results were analyzed by one-way analysis of variance (ANOVA).
Complementary DNA synthesis of TiCRT.
Since the sequence of TiCRT was unknown, we designed the RACEnest1rev, RACEnest2rev, and CRTfw primers (Table 1) based on an expressed sequence tag from T. infestans salivary glands (GenBank: ES597821.1). Total RNA from an insect macerate was extracted with the RNeasy® Mini Kit (Qiagen) and subjected to a reverse transcription polymerase chain reaction (RT-PCR), using the Abridged Anchor Primer and the RACEnest1rev primers (10 μM) (Table 1) with the 5′-RACE® System for Rapid Amplification of cDNA Ends, version 2.0 Kit (Invitrogen, Waltham, MA), according to the manufacturer's instructions. Every primer used in this study was synthesized by Integrated DNA Technologies Inc (San Diego, CA) and is listed in Table 1. The RT-PCR protocol was developed as follows: initial denaturation of 2 minutes at 94°C, 40 cycles each of 45 seconds at 94°C, annealing of 45 seconds at 60°C and extension of 2 minutes at 72°C, and a final extension of 10 minutes at 72°C. An oligo-dC tail was added to the obtained PCR product and this was visualized in an agarose gel stained with ethidium bromide (EthBr) in Tris-borate-EDTA buffer. The band with the expected weight (approximately 1,200 base pairs [bp]) was cut, purified using the Wizard®SV Gel and PCR Clean-Up System Kit (Promega, Madison, WI), and sequenced with the RACEnest1rev primer (Table 1) (Automatic DNA Sequencing Center, Pontificia Universidad Católica de Chile, Santiago, Chile).
Table 1.
Primers used in the cloning of TiCRT and its S domain
| Primer name | Sequence (5′→3′) | Orientation | Use |
|---|---|---|---|
| RACEnest1REV | GAGTTCGTCGTGTTCGTCTCCTGTTTCTGG | Reverse | 5′-RACE |
| RACEnest2REV | GAGTTCGTCTGTTTCGTCTCCTGTTTCTGG | Reverse | 5′-RACE |
| CRTfor | CGGCACAATATTTGGCAACATGCTGATAAC | Forward | 5′-RACE |
| CRTforATG | ATGTGGGCAACAGTATTAAGTT | Forward | 5′-RACE |
| AAP | GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG | Forward | 5′-RACE |
| pQE_fw_SphI | AAAAAAGCATGCATGTGGGCAACAGTATTAAGTT | Forward | TiCRT subcloning |
| pQE_rev_XmaI | TGTCCTCTGCTTGTGCTGCTTGAGGGGCCCAAAAAA | Reverse | TiCRT subcloning |
| TiS_pQE_fw | AAAAAGCATGCATTAATAAAGATATTAGATGCAAGG | Forward | TiS subcloning |
| TiS_pQE_rev | AAAAAGTCGACATTCCATGGTCCTTTATATTTTG | Reverse | TiS subcloning |
TiCRT cloning.
The purified PCR product was ligated into the pGEM®-T Easy vector (Promega), according to the manufacturer's instructions. The recombinant vector pGEM-T/TiCRT was transformed by electroporation into competent Escherichia coli DH5α cells, plated on Luria Bertani agar/ampicillin with 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal) and isopropyl-β-D-1-tiogalactopyranoside (IPTG), and cultivated overnight at 37°C. The scrapped colony was amplified by PCR, under standard proceedings, with the CRTforATG and RACEnest2rev primers (10 μM) (Table 1). The product was purified with the Minipreps DNA Purification System Kit (Promega) and sequenced with T7 promoter and SP6 primers. The sequences obtained were analyzed using the BLAST tool from the National Center for Biotechnology Information (NCBI, MD) database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The TiCRT open reading frame (ORF) was obtained with the tool NCBI ORF Finder (http://www.ncbi.nlm.nih.gov/gorf.html). The TiCRT theoretical molecular weight and isoelectric point were predicted using the ExPASy Computer pI/Mw Tool from the ExPASy Bioinformatics Resources Portal (http://web.expasy.org/compute_pi/). The potential N-glycosylation sites were analyzed with the NetNGlyc 1.0 Server Program (http://www.cbs.dtu.dk/services/NetNGlyc/). The modeling of the tertiary structure of TiCRT was obtained from the primary amino acid structure using the Robetta Beta Full-chain Protein Structure Prediction Server (http://www.robetta.bakerlab.org) and depicted with the UCSF Chimera package (http://www.cgl.ucsf.edu/chimera).
TiCRT subcloning.
For TiCRT subcloning, we used the pQE80L vector (Qiagen), containing six histidine residues at the N-terminal end. A PCR amplification of the TiCRT coding sequence was performed, using Platinum® Taq DNA Polymerase (Invitrogen) and pQE_fw_SphI and pQE_rev_XmaI primers (Table 1). The amplification was carried out according to the following protocol: 2 minutes at 94°C, 40 cycles each of 45 seconds at 94°C, 45 seconds at 62°C, and 2 minutes at 72°C, followed by 10 minutes at 72°C. After double digestion with Sph I and Xma I, the purified TiCRT DNA was inserted into the pQE80L vector also digested, according to the T4 DNA ligase (New England BioLabs, Ipswich, MA) manufacturer's instructions. The recombinant vector pQE80L/TiCRT was transformed by electroporation into competent E. coli BL21(DE3) cells (Novagen, Madison, WI), plated in LB agar/ampicillin and cultured overnight at 37°C. The plasmids were purified and sequenced.
TiCRT expression.
A BL21(DE3)/pQE80L/TiCRT recombinant clone was selected, grown in LB medium with 100 mg/mL ampicillin and incubated in agitation at 37°C overnight. This was added to 250 mL LB medium with ampicillin and grown until an OD600 of 0.5–0.6. Then, IPTG 1 mM was added and incubated in agitation for three additional hours. The culture was centrifuged for 20 minutes at 2,000 × g at 4°C, the supernatant was discarded, and a sample of the pellet was resuspended in 1× PBS and analyzed through denaturating 10% sodium dodecyl sulfate polyacrilamyde gel electrophoresis and Western Blot, under standard conditions.
Sequence analysis and alignment of TiCRT.
The TiCRT nucleotide sequence was translated with the ExPASy Translate Tool from the ExPASy Bioinformatics Resources Portal (http://web.expasy.org/translate/), and the sequence with the wider ORF was selected. The same procedure was performed with the sequence of CRT from human (NCBI Reference Sequence: NM_004343.3), Mus musculus (NCBI Reference Sequence: NM_007591.3), R. prolixus (GenBank: FJ196458.1), Drosophila melanogaster (GenBank: AB000718.1), Apis mellifera (NCBI Reference Sequence: XM_006559506.1), gambiae (GenBank: AF457551.1), and T. cruzi (GenBank: AF457551.1). Afterward, the amino acid sequences were aligned with the Clustal W Program (http://www.ebi.ac.uk/Tools/msa/clustalw2/). A neighbor-joining phylogenetic tree was constructed using the PAUP* 4.b10 Program (http://paup.csit.fsu.edu).
Cloning of the TiCRT S domain.
The coding fragment of TiS (463–861 bp from TiCRT, Figure 3) was amplified by PCR using as template TiCRT DNA, Platinum® Taq DNA Polymerase and the TiS_pQE_fw and TiS_pQE_rev primers (Table 1), and was designed based on the S domains of human CRT (HuCRT) and T. cruzi CRT (TcCRT). Both recombinant proteins are routinely expressed in our laboratory, using conventional procedures. The PCR protocol was developed according to the 5′-rapid amplification of cDNA ends Kit instructions, and consisted of the following series of incubations: 2 minutes at 94°C, 40 cycles each of 45 seconds at 94°C, 45 seconds at 55°C, and 2 minutes at 72°C, followed by 10 minutes at 72°C. The PCR product was visualized in an agarose gel stained with EthBr.
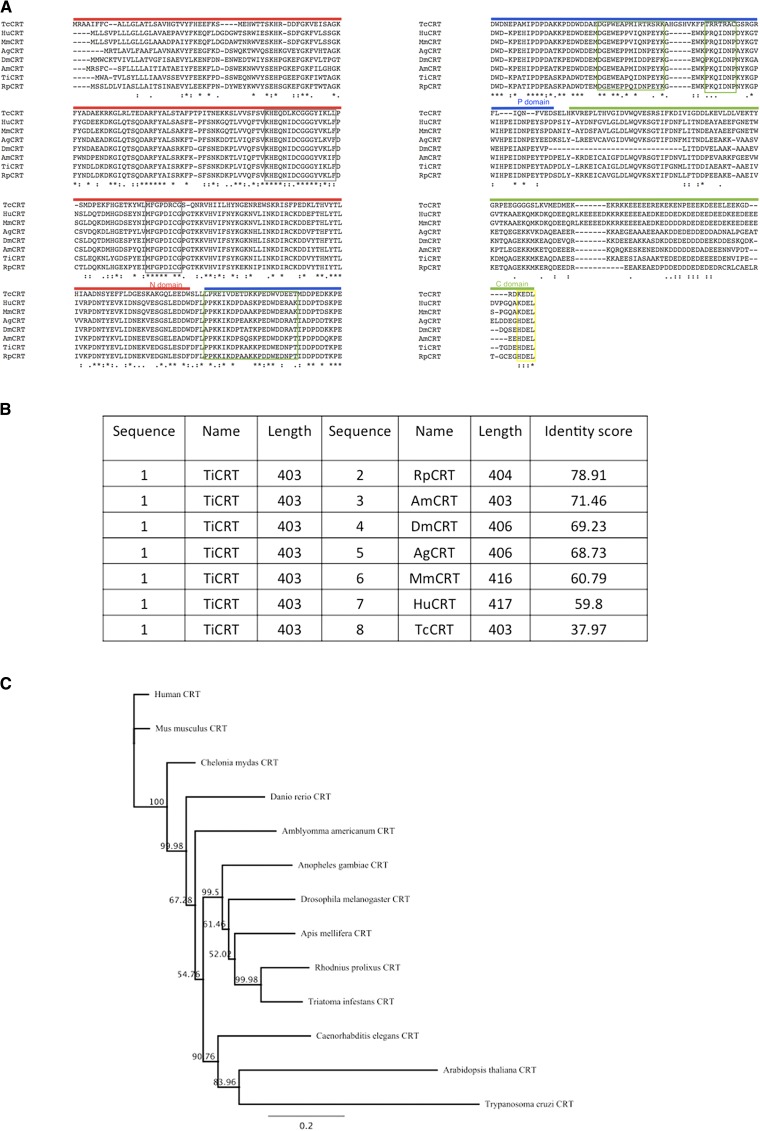
Figure 3.
The TiCRT deduced protein sequence is conserved among species. (A) Alignment of the TiCRT sequence with MmCRT, HuCRT, RpCRT, AmCRT, DmCRT, AgCRT, and TcCRT. The predicted N, P, and C domains are respectively indicated with red, blue, and green lines above the sequence. In both grey boxes, the two characteristic sequences of the CRT N domain (CRT-1 y CRT-2) are shown; in green boxes, the three repeated motifs of the P domain are displayed; and in yellow box, the endoplasmic reticulum (ER) retention signals HDEL or KDEL, are indicated. Conserved residues (*), residues belonging to a same group with strongly conserved properties (:), and residues belonging to a same group with weakly conserved properties (.) are shown. (B) Score table showing sequence identity. (C) Neighbor-joining tree showing the phylogenetic relationships of CRT amino acid sequences, constructed using the PAUP* 4.b10 Program. Sequences of CRT from Amblyomma americanum (GenBank: AAC79094.1), Arabidopsis thaliana (GenBank: AAC49697.1), Danio rerio (GenBank: AAF13700.1), Chelonia mydas (GenBank: EMP37245.1), and Caenorhabditis elegans (NCBI Reference Sequence: NP_504575.1) were added to represent other kingdom and classes. Numbers on the branches are bootstrap values of 5,000 replications.
Subcloning of the TiCRT S domain.
The obtained PCR product was extracted from the agarose gel, purified, and ligated into the pQE80L vector. The construct was transformed by electroporation into competent E. coli BL21(DE3) cells, plated on LB agar/ampicillin and cultured overnight at 37°C. A colony PCR was performed to select the recombinant clones that carried TiS.
Expression of the TiCRT S domain.
The recombinant clone that carried TiS was inoculated into LB with ampicillin and incubated in agitation at 37°C overnight. The culture was poured on 250 mL LB with ampicillin and incubated in shaker at 200 rpm and 37°C until the OD600 reached 0.5–0.6. Then, IPTG 1 mM was added and growing was continued at 150 rpm and 30°C overnight. The next day, the cells were centrifuged for 20 minutes at 2,000 × g and 4°C. The supernatant was discarded and the pellet was resuspended in 30 mL of Tris-HCl, NaCl 500 mM at pH 8.0, imidazole 5 mM, 0.1% Triton X-100, and a protease inhibitor (Cocktail Set V, EDTA-free; Calbiochem, Darmstadt, Germany). The sample was sonicated four times on ice for 30 seconds and centrifuged for 12 minutes at 15,600 × g and 4°C. The pellet was discarded and the supernatant was filtered, purified, and a sample was analyzed by denaturating 10% SDS-PAGE. The filtered supernatant was passed through a Sepharose column charged with Ni2+ 400 mM and eluted with an imidazole gradient. The column was equilibrated with binding buffer (Tris-HCl 160 mM at pH 8.0, NaCl 4 M, and imidazole 40 mM) and the proteins were eluted in a gradient of Tris 80 mM at pH 8.0, NaCl 4 M, and imidazole 4 M. The fractions containing the protein were dialyzed against 1× PBS, analyzed by denaturating SDS-PAGE and Western Blot, using a mouse anti-polyhistidine antibody (Sigma-Aldrich, Saint Louis, MO), followed by a goat alkaline phosphatase–conjugated anti-mouse IgG antibody. Protein concentrations were determined with the Bio-Rad Protein Assay Kit (BioRad, Hercules, CA), according to the manufacturer's indications.
Binding of human C1 to the TiCRT S domain.
For simplicity, the complement reagent used will be designed herein as C1. Indeed, the pure reagent obtained from Complement Technology Inc. (Tyler, TX) corresponds to C1q. This is the collectin, devoid of the C1r and C1s serine proteases. By Western Blot, rTiS (12 μg) was incubated with human C1 (100 μg/mL) and its binding was detected with a goat anti-human C1 antibody followed by a rabbit alkaline phosphatase–conjugated anti-goat IgG antibody. As positive controls, human C1 (200 ng) was detected with a goat anti-human C1q antibody, followed by a rabbit alkaline phosphatase–conjugated anti-goat IgG antibody. We also analyzed the S domain of TcCRT (TcS), previously obtained in our laboratory.19 TcS was detected with a rabbit anti-TcS polyclonal antibody (previously generated in our laboratory19), followed by a goat alkaline phosphatase–conjugated anti-rabbit IgG antibody. As negative control, TiS (12 μg) was incubated with 3% PBS/skimmed milk, followed by a goat anti-human C1 antibody and a rabbit alkaline phosphatase–conjugated anti-goat IgG antibody. They were all revealed with 4-nitroblue tetrazolium chloride solution and 5-bromo-4-chloro-3-indoyl-phosphate. Binding of C1 to rTiS was also demonstrated by Dot Blot, as shown in Supplemental Figure 1.
Inhibition of C4b deposit by the TiCRT S domain.
Microtitration plates were coated with 100 μL of human IgM at 4 μg/mL and incubated at 4°C overnight. Nonspecific binding sites were blocked with 250 μL of 2.5% w/v casein in PBS, and the following was added: 1) 100 μL of casein; 2) 100 μL of rTcCRT; 3) 100 μL of rHuCRT; 4) 100 μL of EDTA 10 mM; 5) 100 μL of rTcS; and 6) 100 μL of rTiS. In 1–6, the reagents were added at 2.4, 4.8, and 9.6 μM and were diluted in human normal serum (HNS) as a source of complement human C4. Casein was the negative inhibition control, whereas EDTA was the positive one. They were all incubated for 1 hour at 37°C in Veronal Buffer. C4b deposits were detected with an affinity-purified rabbit anti-human C4 antibody, followed by an affinity-purified goat HRP-conjugated anti-rabbit IgG antibody. HRP activity (measured at 405 nm) was assessed by addition of ABTS with 0.03% v/v H2O2. The results were analyzed by two-way ANOVA.
Results
No activation of the classical pathway of complement is detected in blood recovered from the anterior midgut of T. infestans.
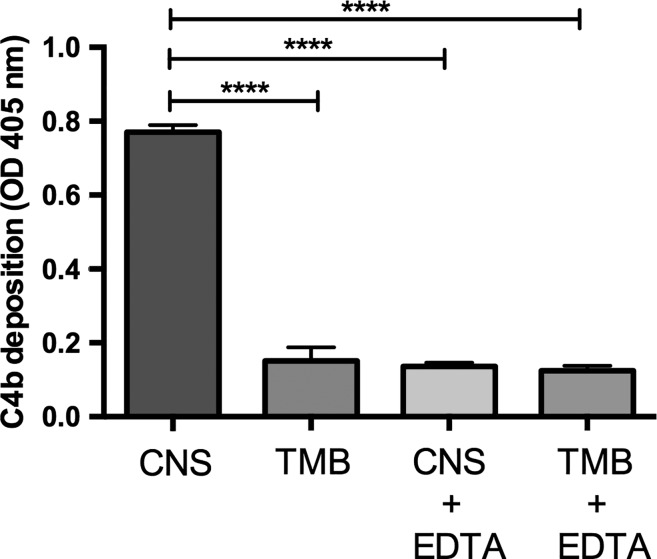
As shown in Figure 1 , a drastic decrease in the C4b deposition of the blood recovered from T. infestans anterior midgut was observed as compared with the C4b deposition of CNS. This result indicates that in the insect's anterior midgut conditions are such that the classical pathway of the complement system is importantly inactivated. Therefore, the possibility stands that, in agreement with our hypothesis, and as a simple proof-of-concept procedure, salivary CRT may be at least partially involved in such inactivation. We are aware that this experiment will not probe that salivary TiCRT is indeed responsible for complement inactivation. However, should the result be negative (no complement inactivation), it could not be argued that salivary TiCRT is involved.
Figure 1.
C4b deposition is inhibited in blood recovered from the Triatoma infestans anterior midgut. Plates were sensitized with IgM and C4b deposition (a direct reflection of C4 activation) was measured in blood recovered from the T. infestans anterior midgut (TMB) immediately after feeding. This C4b deposit was compared with chicken normal serum (CNS) obtained from the same bird used to feed the insects. CNS plus ethylenediaminetetraacetic acid (CNS + EDTA) and TMB plus EDTA (TMB + EDTA) were positive inhibition controls. The results correspond to the mean of triplicates and were analyzed by one-way analysis of variance (P < 0.0001) using the GraphPad Prism Program version 6.0c.
TiCRT DNA coding sequence identification.
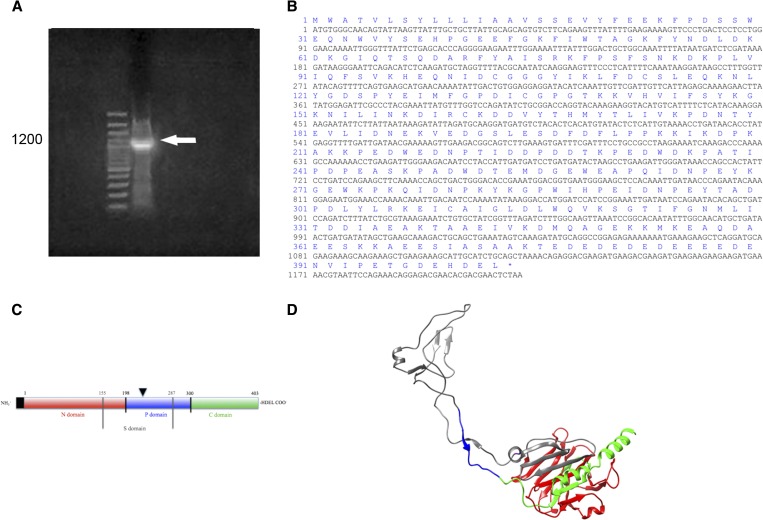
The first reported TiCRT partial sequence obtained by 5′-RACE, included a segment of 400 bp containing the C-terminal domain, the stop codon and the endoplasmic reticulum (ER) retention signal, HDEL. Based on this, the full-length sequence of the TiCRT DNA coding sequence was obtained. This information was also useful for designing the CRTforATG and RACEnest2rev primers (Table 1), since they amplified the complete TiCRT ORF (Figure 2A ). The TiCRT ORF presented 1,212 bp, with a predicted 403-residue amino acid sequence. As shown in Figure 2B, the nucleotide sequence presented the ATG initiation and the TAA stop codons. As expected, we observed the initial methionine and the ER retention signal HDEL. The theoretical molecular weight of TiCRT was 46.17 kDa and the isoelectric point 4.36. In the schematic diagram of the TiCRT deduced protein in Figure 2C, the protein domains and lengths are shown. A three-dimensional model of TiCRT is shown in Figure 2D.
Figure 2.
TiCRT DNA coding sequence identification. (A) 1% agarose gel. Lane 1: 10,000–base pair (bp) ladder; lane 2: nucleotide TiCRT 5′-RACE amplification product of 1,200 bp. (B) Nucleotide and predicted amino acid sequence of TiCRT. (C) Simplified characterization of the TiCRT deduced protein. The N, P, and C domains, residues 1–198, 199–300, and 301–403, are respectively shown. TiS (residues 155–287) is shown between grey lines. The leader sequence is indicated in a black box and the endoplasmic reticulum (ER) retention signal HDEL is shown at the C-terminal end. The glycosylation site is shown with an inverted black triangle (N220). (D) Predicted three-dimensional model of the TiCRT deduced protein. Globular N, extended P, S (grey), and C domains, the last one ending in an α-helix, are shown. The color code of Figure 2C is also valid for Figure 2D. It presents a QMEAN4 of −0.41, indicating that the three-dimensional TiCRT model obtained is within the accepted range of closeness to the protein native conformation (−1 to 1).
TiCRT shares important primary structure with insect and mammal CRTs, but not with TcCRT.
Figure 3A shows the alignment of TiCRT with different CRTs. In all these sequences, the N, P, and C domains, as well as the ER retention signal HDEL (in TiCRT, RpCRT, AmCRT, DmCRT, and AgCRT) or KDEL (in HuCRT, MmCRT, and TcCRT), are detectable. The HDEL retention signal seems to identify insect CRTs. The two characteristic N domain sequences of the CRT-1 and CRT-2 families, KHEQNIDCGGGYIKLF (residues 96–111) and MFGPDICG (residues 129–136); besides the three P domain repeated motifs, PPKKIKDPKAKKPED (residues 202–222), GEWEAPQIDNPEYK (residues 257–270), and PKQIDNP (residues 275–281), are shown (Figure 3A). As shown in Figure 3B, the TiCRT amino acid sequence presents variable identity with CRTs from several species: 78.9% with R. prolixus (RpCRT), 71.5% with A. mellifera (AmCRT), 69.2% with D. melanogaster (DMCRT), 68.7% with A. gambiae (AgCRT), 60.8% with M. musculus (MmCRT), 59.8% with human (HuCRT), and 38% with T. cruzi (TcCRT). This indicates that CRT protein sequences are conserved among species. A neighbor-joining phylogenetic tree showing the CRT relationships in different species is also shown (Figure 3C).
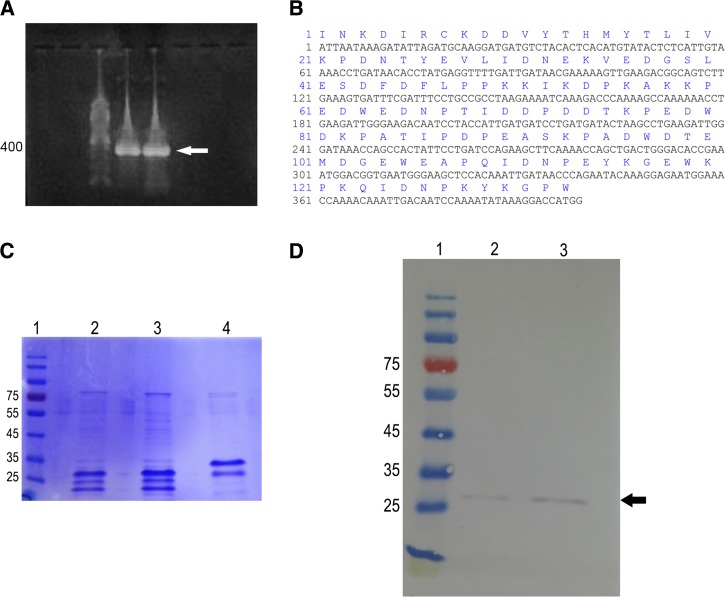
Recombinant TiCRT S domain expression and purification.
As expected, in Figure 4A , we observed the 399-bp band corresponding to the PCR-amplified fragment of TiS. The nucleotide and amino acid sequence of TiS is shown in Figure 4B. Figure 4C shows the denaturating 10% SDS-PAGE patterns of TiS and TcS, that display 30 and 34 kDa, respectively. The expression of TiS was also analyzed by Western Blot using a mouse anti-polyhistidine antibody (Figure 4D).
Figure 4.
TiCRT S domain coding sequence identification and recombinant protein identification. (A) TiS nucleotide sequence amplification (1% agarose gel). Lane 1: 100-bp ladder; lanes 2 and 3: TiS polymerase chain reaction (PCR) expected product of 399 bp. (B) TiS nucleotide and deduced amino acid sequence with 399 bp and 133 residues, respectively, are shown. (C) TiS peptide expression (10% denaturant SDS-PAGE). Lane 1: Molecular weight marker; lanes 2 and 3: 6 and 12 μg of purified recombinant TiS, respectively; lane 4: purified recombinant TcS (6 μg). (D) Identification of purified TiS by Western Blot. Lane 1: Molecular weight marker; lanes 2 and 3: 40 and 80 ng of purified recombinant TiS, respectively, detected with a mouse anti-polyhistidine antibody, followed by a goat alkaline phosphatase–conjugated anti-mouse IgG antibody.
C1 binds to the TiCRT S domain.
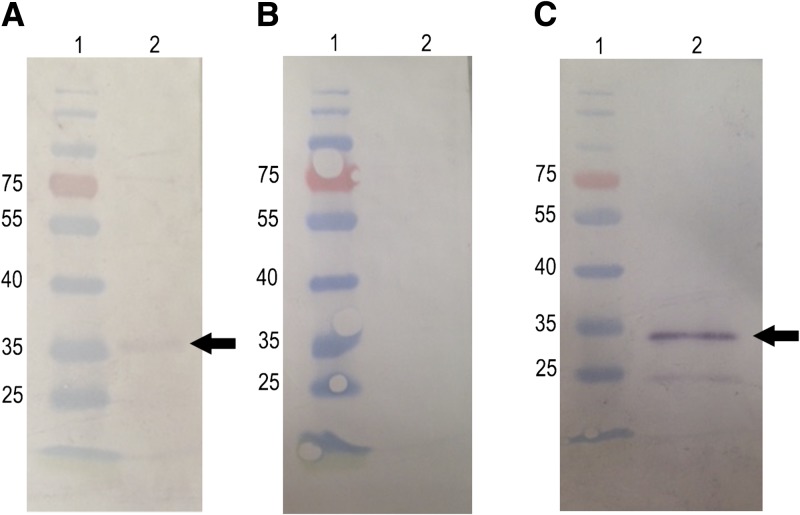
CRT from diverse origins binds C1, interfering with the function of this molecule.21–25 By Western Blot, we showed that rTiS also binds C1, as detected by an anti-C1 antibody (Figures 5A and Supplemental Figure 1).
Figure 5.
Human C1 binds to the TiCRT recombinant S domain. (A) rTiS was incubated with human C1, followed by a goat anti-C1 antibody and a rabbit alkaline phosphatase–conjugated anti-goat IgG antibody. (B) As a negative control, rTiS was incubated with 3% phosphate-buffered saline (PBS)/skimmed milk, followed by goat anti-C1 antibody and a rabbit alkaline phosphatase–conjugated anti-goat IgG antibody. (C) As a positive control, C1 was detected with an anti-C1 antibody, followed by a rabbit alkaline phosphatase–conjugated anti-goat IgG antibody.
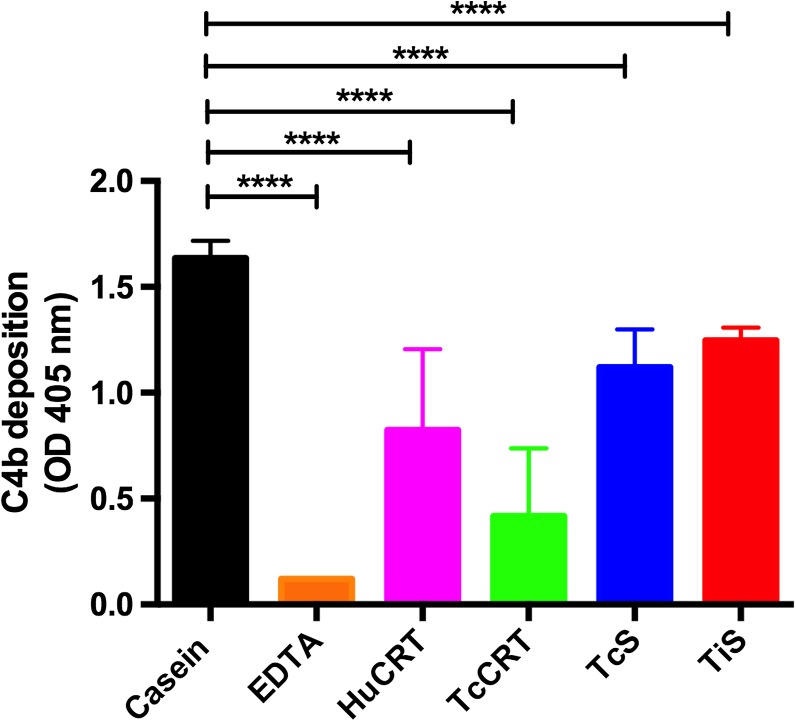
The TiCRT S domain inhibits the classical pathway of complement.
As shown in Figure 6 , TiS inhibited the deposit of C4b. Casein, a protein that has no effect on the complement system, was used as a negative control. EDTA, a Ca2+ chelator, was the positive inhibition control. HuCRT, TcCRT, and TcS were included also as positive inhibition controls, given their known capacity to inhibit the classical pathway of complement. We observed that TiS also inhibits this pathway of complement, although to a lower grade than complete proteins (HuCRT and TcCRT). TcS showed an inhibition degree similar to that of TiS in HNS.
Figure 6.
TiS inhibits the classical pathway of the complement system. Microtitration plates were sensitized with IgM, and rTiS was added together with human normal serum (HNS), as a source of complement components C1 and C4. As negative and positive inhibition controls, casein and EDTA were respectively used. HuCRT, TcCRT, and TcS were also tested. The results correspond to the mean of triplicates and were analyzed by two-way analysis of variance and Dunnett's multiple comparison test, casein being the control group (P < 0.0001), using the GraphPad Prism Program version 6.0c.
Discussion
Triatominae insects ingest about ten times their body weight in their blood meals.26–28 The complement system, mainly present in blood, has at least four macromolecules specialized in the detection of an impressive variety of molecular danger signals. The system can also be activated by default, when it detects the lack of molecular structures normally present in vertebrates, as is the case of the alternative pathway. Thus, membrane-destructive, opsonizing, immune stimulatory, and pro-inflammatory activities are generated.14 It is conceivable that, given its molecular complexity, the insect's digestive tract may activate the complement system, even in the absence of antibodies. Accordingly, the results presented herein are consistent with the possibility that CRT present in T. infestans saliva, by virtue of its capacity to inhibit complement activation, can counteract the complement potential deleterious effects on the digestive tissues in these insects.
Triatoma infestans possesses in its stylet two channels (salivary and alimentary) with afferent and efferent respective uses, connected at the anterior end.29 It is then possible that saliva, ejected at the bite site, contains molecules that inhibit the complement proteins present in the massive blood meal that floods the first part of the insect's digestive tract. We propose that CRT, a multifunctional molecule, known to be present in salivary gland extracts from insects30,31 and to inhibit the complement system,19,32,33 could be responsible at least partly, for this effect (A dynamic view of this functional perception is presented in Supplemental Video).
We have investigated the complement functional state of the blood contained in the anterior midgut of T. infestans immediately after feeding. Complement activity, measured by the C4b deposition, was drastically inhibited in the anterior midgut of the insect, as compared with CNS from the same bird used for feeding purposes (Figure 1). These results are compatible with those of Khattab and others that found C3a and C5b-C9 in the midgut of Anopheles stephensi as indicators of complement activation, mainly via the alternative pathway.34 Our results, by measuring C4b in blood retrieved from the anterior midgut of T. infestans, indicate an inhibitory effect on the classical pathway. Whether both protective strategies are operative in T. infestans, remains to be determined.
The presence of TiCRT in the T. infestans salivary glands, together with a potent C4b inhibition in the anterior midgut of the insect, withstands our hypothesis proposing that the salivary chaperone molecule is involved in this complement inhibition. It was thus necessary to attempt the cloning and characterization of the full-length coding sequence DNA of the TiCRT gene.
Our results showed that the coding sequence of the TiCRT gene spans 403 amino acids (Figures 2A–C). The modeling of the tertiary structure of TiCRT showed the globular N domain and the central P domain with its characteristic loop (Figure 2D). The calculated theoretical molecular weight of TiCRT was 46 kDa and the isoelectric point 4.4.
The deduced amino acid sequence contained several features common to all CRTs and its three-major domains15 (N-terminal, central P, and C-terminal). The two potential CRT family signature motifs KHEQNIDCGGGYIKLF and MFGPDICG, which are highly conserved among CRTs, were found in the TiCRT N domain. The P domain included the three repeats CRT motifs, PPKKIKDPKAKKPED, GEWEAPQIDNPEYK, and PKQIDNP.
In agreement with the sequence of R. prolixus, A. mellifera, D. melanogaster, and A. gambiae, the ER retention sequence HDEL was present at the end of the C-terminal domain (Figure 3A). Alignment of the TiCRT amino acid sequence with R. prolixus CRT revealed 79% and approximately 70% identities with CRT from other insects. With mammals, CRT identity was about 60%, whereas with TcCRT, it was 38% (Figure 3B). The relationship of TiCRT is much closer with CRT from insects than with fish, reptile, and mammal CRTs, and the more distant relation is with TcCRT. Surprisingly, the neighbor-joining tree showed that TcCRT is closer to CRT from plants than any other CRT (Figure 3C).
Similar to those of some higher plants and insects,35 in TiCRT, the ER retrieval signal was HDEL, instead of the typical KDEL found in some mammals and parasites. In spite of the presence of ER retention sequences in CRT, in some parasites, the protein is released from the cells. The missing KDEL sequence in CRT from ticks, which is secreted in saliva, could contribute to its routing into a secretory pathway rather than being retained in the ER.30 In support of this theory, the tick Amblyomma americanum, while feeding in their host, secretes CRT, presumably as a mechanism to divert host defensive responses.36 Moreover, CRTs from several ticks have an ER retrieval signal HEEL,30,37 whereas two CRTs from Caenorhabditis elegans have no ER retrieval signal.38 These investigations indicate that CRTs from closely related species will likely display the same signal. The biological significance of these differences is unknown.
CRT binds to C1, as shown for HuCRT21 and CRTs from Necator americanus,22 Haemonchus contortus,23 Trypanosoma carasii,24 T. cruzi,25 Entamoeba histolytica, and Entamoeba dispar.32 Resulting inhibition of the classical pathway of the complement system is detectable.19,32,33,39 Based on this rationale, we asked whether this capacity was present in TiCRT.
Perhaps, poor expression caused by toxicity in the host cell, insolubility, mRNA secondary structure that prevents interactions with cellular machinery, or uncontrolled host cell basal protein expression inhibited the recombinant protein yield, thus explaining our impossibility to express TiCRT.
Given the known relevance of the S domain from other species in complement inhibition,18 we cloned, sequenced, expressed, and purified the TiCRT S domain (Figures 4A–D). As expected, this domain binds to human C1 (Figures 5A and Supplemental Figure 1) and inhibits the classical pathway of the complement system in HNS (Figure 5A). It is not surprising that the TcS and TiS domains show a lesser inhibitory activity as compared with complete TcCRT and HuCRT, given that this and other functions involve several domains correctly folded in the intact chaperone molecule.
In synthesis, we have:1) identified the entire nucleotide coding sequence of the TiCRT gene; 2) deduced the full-length sequence of the protein; 3) established its phylogenetic homology to other CRTs; 4) cloned, sequenced, and expressed the TiCRT S domain; 5) determined the TiS capacity to bind complement component C1 and to inhibit the classical pathway of complement; and 6) proposed that salivary TiCRT could be responsible, at least partly, for the protection of the insect's digestive tract against the damage of complement proteins present in the vertebrate blood. In this scenery, recombinant proteins or relevant domains from hematophagous salivary origins, such as CRT, could be part of vaccines similar to those against the Bm86 polypeptide “hidden antigen” from ticks, used to immunize cattle.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ruth Mora, Nancy Fabres, and Antonio Rojas for expert technical support, and Gonzalo Cabrera, Norbel Galanti, and Aldo Solari for guidance and critical analysis of the data.
Footnotes
Authors' addresses: Katherine Weinberger, Norberto Collazo, Juan Carlos Aguillón, María Carmen Molina, Carlos Rosas, Jaime Peña, Javier Pizarro, Ismael Maldonado, and Arturo Ferreira, Program of Immunology, Institute of Biomedical Sciences, Faculty of Medicine, Universidad de Chile, Santiago, Chile, E-mails: kta.weinberger@gmail.com, norberto.collazo@gmail.com, jaguillo@med.uchile.cl, mcmolina@med.uchile.cl, carlosrosas@u.uchile.cl, jaime.pena.alvarez@gmail.com, jpizarrob@ug.uchile.cl, ismael_amf@yahoo.es, and aferreir@med.uchile.cl. Pedro E. Cattan, Departamento de Ciencias Biológicas Animales, Universidad de Chile, Santiago, Chile, E-mail: pcattan@u.uchile.cl. Werner Apt, Clinical Investigation, North Unit of Parasitology, Faculty of Medicine, University of Chile, Santiago, Chile, E-mail: wapt@med.uchile.cl.
References
- 1.Canals M, Ehrenfeld M, Cattan P. Situation of Mepraia spinolai, a wild vector for Chagas disease in Chile, in relation to other vector from the perspective of their feeding profile. Rev Med Chil. 2000;128:1108–1112. [PubMed] [Google Scholar]
- 2.González CR, Reyes C, Canals A, Parra A, Muñoz X, Rodríguez K. An entomological and seroepidemiological study of the vectorial-transmission risk of Chagas disease in the coast of northern Chile. Med Vet Entomol. 2015;29:387–392. doi: 10.1111/mve.12131. [DOI] [PubMed] [Google Scholar]
- 3.Bern C. Chagas' disease. N Engl J Med. 2015;373:456–466. doi: 10.1056/NEJMra1410150. [DOI] [PubMed] [Google Scholar]
- 4.Hotez PJ, Dumonteil E, Woc-Colburn L, Serpa JA, Bezek S, Edwards MS, Hallmark CJ, Musselwhite LW, Flink BJ, Bottazzi ME. Chagas disease: “the new HIV/AIDS of the Americas. PLoS Negl Trop Dis. 2012;6:e1498. doi: 10.1371/journal.pntd.0001498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodrigues J, Albajar P. Chagas disease: a new worldwide challenge. Nature. 2010;465:S6–S7. doi: 10.1038/nature09221. [DOI] [PubMed] [Google Scholar]
- 6.Andrade BB, Teixeira CR, Barral A, Barral-Netto M. Haematophagous arthropod saliva and host defence system: a tale of tear and blood. An Acad Bras Cienc. 2005;77:665–693. doi: 10.1590/s0001-37652005000400008. [DOI] [PubMed] [Google Scholar]
- 7.Champagne DE. The role of salivary vasodilators in bloodfeeding and parasite transmission. Parasitol Today. 1994;10:430–433. doi: 10.1016/0169-4758(94)90173-2. [DOI] [PubMed] [Google Scholar]
- 8.Ribeiro JM. Blood-feeding arthropods: live syringes or invertebrate pharmacologists? Infect Agents Dis. 1995;4:143–152. [PubMed] [Google Scholar]
- 9.Stark KR, James AA. Salivary gland anticoagulants in culicine and anopheline mosquitoes (Diptera: Culicidae) J Med Entomol. 1996;33:645–650. doi: 10.1093/jmedent/33.4.645. [DOI] [PubMed] [Google Scholar]
- 10.Ribeiro JMC, Francischetti IMB. Role of arthropod saliva in blood feeding: sialome and post-sialome perspectives. Annu Rev Entomol. 2003;48:73–88. doi: 10.1146/annurev.ento.48.060402.102812. [DOI] [PubMed] [Google Scholar]
- 11.Fontaine A, Diouf I, Bakkali N, Missé D, Pagès F, Fusai T, Roger C, Almeras L. Implication of haematophagous arthropod salivary proteins in host-vector interactions. Parasit Vectors. 2011;4:187. doi: 10.1186/1756-3305-4-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barros VC, Assumpçao JC, Cadete AM, Santos VC, Cavalcante RR, Araújo RN, Pereria MH, Gontijo NF. The role of salivary and intestinal complement system inhibitors in the midgut protection of triatomines and mosquitoes. PLoS One. 2009;4:e6047. doi: 10.1371/journal.pone.0006047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janeway CA, Jr, Travers P, Walport M, Schlomchik MJ. 5th edition. New York, NY: Garland Science; 2001. The complement system and innate immunity. Immunobiology: The Immune System in Health and Disease. [Google Scholar]
- 15.Michalak M, Corbett EF, Mesaeli N, Nakamura K, Opas M. Calreticulin: one protein, one gene, many functions. Biochem J. 1999;344:281–292. [PMC free article] [PubMed] [Google Scholar]
- 16.Coppolino MG, Dedhar S. Calreticulin. Int J Biochem Cell Biol. 1998;30:553–558. doi: 10.1016/s1357-2725(97)00153-2. [DOI] [PubMed] [Google Scholar]
- 17.Baksh S, Michalak M. Expression of calreticulin in Escherichia coli and identification of its Ca2+ binding domains. J Biol Chem. 1991;266:21458–21465. [PubMed] [Google Scholar]
- 18.Lynch NJ, Schneider H, Sim RB, Bickel U, Schwaeble WJ. In vivo pharmacokinetics of calreticulin S-domain, an inhibitor of the classical complement pathway. Int Immunopharmacol. 2002;2:415–422. doi: 10.1016/s1567-5769(01)00165-5. [DOI] [PubMed] [Google Scholar]
- 19.Ferreira V, Valck C, Sánchez G, Gingras A, Tzima S, Molina MC, Sim R, Schwaeble W, Ferreira A. The classical activation pathway of the human complement system is specifically inhibited by calreticulin from Trypanosoma cruzi. J Immunol. 2004;172:3042–3050. doi: 10.4049/jimmunol.172.5.3042. [DOI] [PubMed] [Google Scholar]
- 20.Cavalcante RR, Pereira MH, Gontijo NF. Anti-complement activity in the saliva of phlebotomine sand flies and other haematophagous insects. Parasitol. 2003;127:87–93. doi: 10.1017/s0031182003003329. [DOI] [PubMed] [Google Scholar]
- 21.Malhotra R, Thiel S, Reid KB, Sim RB. Human leukocyte C1q receptor binds other soluble proteins with collagen domains. J Exp Med. 1990;172:955–959. doi: 10.1084/jem.172.3.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kasper G, Brown A, Eberl M, Vallar L, Kieffer N, Berry C, Girdwood K, Eggleton P, Quinnell R, Protchard DI. A calreticulin-like molecule from the human hookworm Necator americanus interacts with C1q and the cytoplasmic signalling domains of some integrins. Parasite Immunol. 2001;23:141–152. doi: 10.1046/j.1365-3024.2001.00366.x. [DOI] [PubMed] [Google Scholar]
- 23.Naresha S, Suryawanshi A, Agarwal M, Singh BP, Joshi P. Mapping the complement C1q binding site in Haemonchus contortus calreticulin. Mol Biochem Parasitol. 2009;166:42–46. doi: 10.1016/j.molbiopara.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Oladiran A, Belosevic M. Trypanosoma carassii calreticulin binds host complement component C1q and inhibits classical complement pathway-mediated lysis. Dev Comp Immunol. 2009;34:396–405. doi: 10.1016/j.dci.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Ramírez G, Valck C, Molina MC, Ribeiro CH, López N, Sánchez G, Ferreira VP, Billeta R, Aguilar L, Maldonado I, Cattán P, Schwaeble W, Ferreira A. Trypanosoma cruzi calreticulin: a novel virulence factor that binds complement C1 on the parasite surface and promotes infectivity. Immunobiol. 2011;216:265–273. doi: 10.1016/j.imbio.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 26.Zeledón R, Guardia VM, Zuñiga A, Swartzwelder JC. Biology and ethology of Triatoma dimidata (Latreille, 1811). Life cycle, amount of blood ingested, resistance of starvation, and size of adults. J Med Entomol. 1970;7:313–319. doi: 10.1093/jmedent/7.3.313. [DOI] [PubMed] [Google Scholar]
- 27.Pietrokovsky S, Bottazi V, Schweigmann N, Hedo A, Wisnivesky-Colli C. Comparison of the blood meal size among Triatoma infestans, T. guasayana and T. sordida (Hemiptera: Reduviidae) of Argentina under laboratory conditions. Mem Inst Qswaldo Cruz. 1996;91:241–242. doi: 10.1590/s0074-02761996000200022. [DOI] [PubMed] [Google Scholar]
- 28.Alzamora A, Correa P, Gaggero E, Acuña-Retamar M, Cattán PE. Feeding behaviour of Mepraia spinolai in two frequent hosts from its habitat. Parasitol Latinoam. 2007;62:112–117. [Google Scholar]
- 29.Krenn HW, Aspök H. Form, function and evolution of the mouthparts of blood-feeding Arthropoda. Arthropod Struct Dev. 2012;41:101–118. doi: 10.1016/j.asd.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Jaworski DC, Simmen FA, Lamoreaux W, Needham GR. A secreted calreticulin protein in ixodid tick (Amblyomma americanum) saliva. J Insect Physiol. 1995;41:369–375. [Google Scholar]
- 31.Jaworski DC, Higgins JA, Radulovic S, Vaughan JJ, Azad AF. Presence of calreticulin in vector fleas (Siphonaptera) J Med Entomol. 1996;33:482–489. doi: 10.1093/jmedent/33.3.482. [DOI] [PubMed] [Google Scholar]
- 32.Ximénez C, González E, Nieves ME, Silva-Olivares A, Shibayama M, Galindo-Gómez S, Escobar-Herrera J, García de León MC, Morán P, Valadez A, Rojas L, Hernández EG, Partida O, Cerritos R. Entamoeba histolytica and Entamoeba dispar calreticulin: inhibition of classical complement pathway and differences in the level of expression in amoebic liver abscess. BioMed Res Int. 2014;2014:127453. doi: 10.1155/2014/127453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sosoniuk E, Vallejos G, Kenawy H, Gaboriaud C, Thielens N, Fujita T, Schwaeble W, Ferreira A, Valck C. Trypanosoma cruzi calreticulin inhibits the complement lectin pathway activation by direct interaction with L-Ficolin. Mol Immunol. 2014;60:80–85. doi: 10.1016/j.molimm.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 34.Khattab A, Barroso M, Miettinen T, Meri S. Anopheles midgut epithelium evades human complement activity by capturing factor H from the blood meal. PLoS Negl Trop Dis. 2015;9:e0003513. doi: 10.1371/journal.pntd.0003513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Opas M, Dziak E, Fliegel L, Michalak M. Regulation of expression and intracellular distribution of calreticulin, a major calcium binding protein of non muscle cells. J Cell Physiol. 1991;149:160–171. doi: 10.1002/jcp.1041490120. [DOI] [PubMed] [Google Scholar]
- 36.Kovacs H, Campbell ID, Strong P, Johnson S, Ward FJ, Reid KB, Eggelton P. Evidence that C1q binds specifically to CH2-like immunoglobulin C motifs present in the autoantigen calreticulin and interferes with complement activation. Biochem. 1998;37:17865–17874. doi: 10.1021/bi973197p. [DOI] [PubMed] [Google Scholar]
- 37.Xu G, Fang QQ, Keirans JE, Durden LA. Cloning and sequencing of putative calreticulin complementary DNAs from four hard tick species. J Parasitol. 2004;90:73–78. doi: 10.1645/GE-157R. [DOI] [PubMed] [Google Scholar]
- 38.Tsuji N, Morales TH, Ozols VV, Carmody AB, Chandrashekar R. Molecular characterization of a calcium-binding protein from the filarial parasite Dirofilaria immitis. Mol Biochem Parasitol. 1998;97:69–79. doi: 10.1016/s0166-6851(98)00131-5. [DOI] [PubMed] [Google Scholar]
- 39.Johnson S, Michalak M, Opas M, Eggleton P. The ins and outs of calreticulin: from the ER lumen to the extracellular space. Trends Cell Biol. 2001;11:122–129. doi: 10.1016/s0962-8924(01)01926-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.