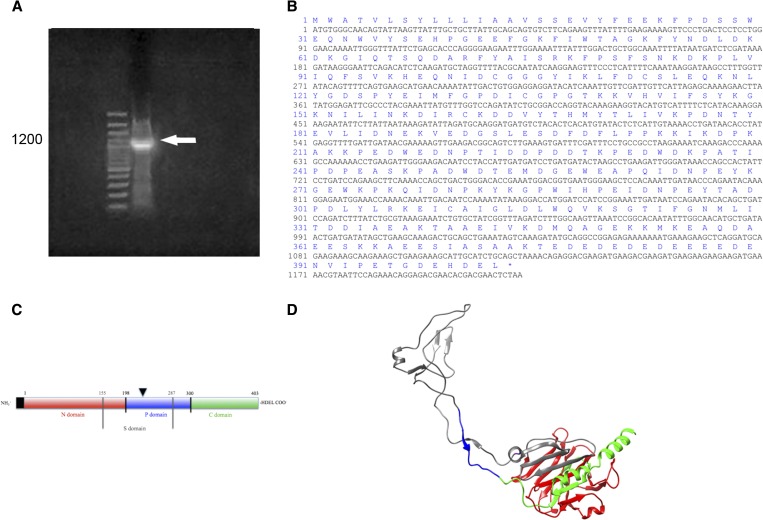
Figure 2.
TiCRT DNA coding sequence identification. (A) 1% agarose gel. Lane 1: 10,000–base pair (bp) ladder; lane 2: nucleotide TiCRT 5′-RACE amplification product of 1,200 bp. (B) Nucleotide and predicted amino acid sequence of TiCRT. (C) Simplified characterization of the TiCRT deduced protein. The N, P, and C domains, residues 1–198, 199–300, and 301–403, are respectively shown. TiS (residues 155–287) is shown between grey lines. The leader sequence is indicated in a black box and the endoplasmic reticulum (ER) retention signal HDEL is shown at the C-terminal end. The glycosylation site is shown with an inverted black triangle (N220). (D) Predicted three-dimensional model of the TiCRT deduced protein. Globular N, extended P, S (grey), and C domains, the last one ending in an α-helix, are shown. The color code of Figure 2C is also valid for Figure 2D. It presents a QMEAN4 of −0.41, indicating that the three-dimensional TiCRT model obtained is within the accepted range of closeness to the protein native conformation (−1 to 1).