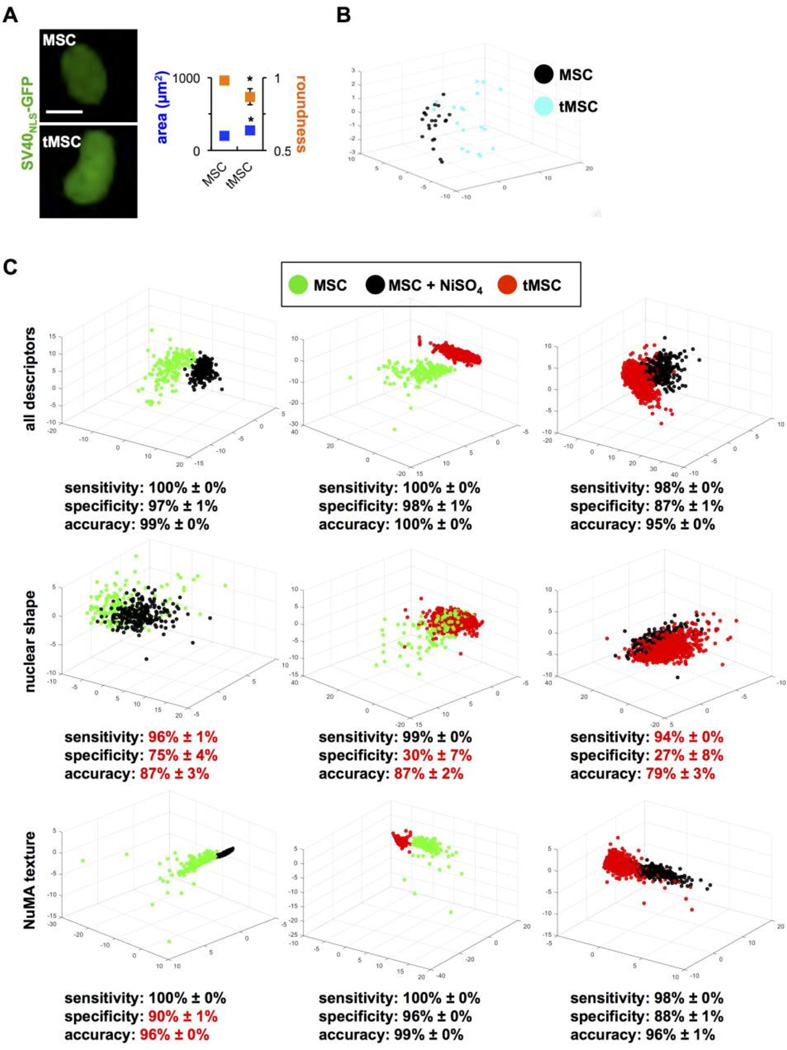
Figure 7. NuMA textural descriptors parse transformed and transforming MSCs.
(A) Classic shape parameters (nuclear size and roundness) computed from GFP signals in normal and transformed MSCs transduced with SV40NLS-GFP to label cell nuclei. *, P < 0.001. (B) Highcontent morphotextural analysis based on NuMA and DNA staining of MSCs, NiSO4-treated MSCs (three days after treatment), and tMSCs. All descriptors were used for the analyses in the top panels, whereas a subset of nuclear shape or NuMA texture descriptors were used for the analyses in the middle and lower panels, respectively. Classifying metrics are shown, with red highlights identifying worse classification compared to the full pool of descriptors.