Abstract
Purpose of review
To provide an overview of the biological processes implicated in chromatin-based pathways that control endothelial gene expression patterns in both health and disease and highlight how these processes are relevant to cardiovascular disease.
Recent findings
Epigenetics refers to chromatin-based pathways important in the regulation of gene expression and includes three distinct, but highly interrelated, mechanisms: DNA methylation, histone density and posttranslational modifications, and RNA-based mechanisms. It is of great interest that epigenetic regulation of genes enriched in the vascular endothelium is a prominent regulatory pathway. How environmental cues within the vasculature, such as hemodynamic forces or hypoxia, influence these epigenetic mechanisms will be reviewed.
Summary
Although a newer area for study, exciting new evidence identifies that epigenetic processes are highly dynamic and respond to a myriad of environmental stimuli. Integrating chromatin-based pathways into our understanding of gene expression offers newer insight into disease processes.
Keywords: cardiovascular disease, DNA methylation, epigenetics, histone posttranslational modifications
Introduction
The chromosomal theory of inheritance, for which Thomas Hunt Morgan won the Nobel Prize in 1933, argued for the critical role of the chromosome in heredity. When first proposed, however, it was not immediately accepted. Such concerns grew with the recognition that the A, C, G, and T nucleotide content of the static genetic DNA was, for the most part, identical in normal diploid cells. How could an endothelial cell, a vascular smooth muscle cell (VSMC), or a cardiac myocyte exhibit a distinct cellular phenotype if the DNA was identical across cell types? Broadly defined, epigenetics refers to chromatin-based mechanisms important in the regulation of gene expression that do not involve changes to the DNA sequence per se.
Epigenetics provides a newer perspective for understanding how gene expression is perturbed in prevalent diseases of the human vascular system characterized by a dysfunctional endothelium. These pathways offer a new perspective on gene regulation that extends the classic cis/trans paradigm and helps to explain some of its limitations. The genomics era has exploited genotype/phenotype associations as they relate to the susceptibility of diseases, especially complex ones like atherosclerosis. These genome-wide association studies (GWASs) have sought to identify genetic determinants of cardiovascular disease. However, two major dilemmas with these studies have emerged: they identified loci which do not correspond to protein-coding genes [1•,2•] and the effects and contribution of environmental factors such as diet, exercise, socioeconomic status, and developmental stresses are ignored [3•]. This review provides a background on epigenetic processes in health and disease, and highlights relevant processes to the development of cardiovascular disease using the endothelial cell as a model cell for discussion.
Epigenetic processes
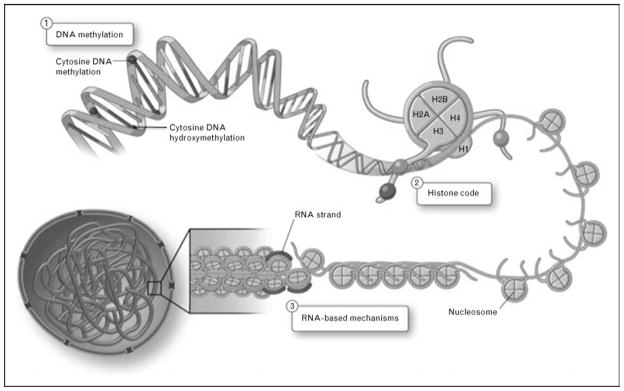
The International Human Epigenome Consortium (IHEC) was launched in January 2010 and reminds us that a greater understanding of epigenetic mechanisms is coming to the forefront. This large-scale project aims to catalogue the epigenetic marks, especially DNA methylation and histone modifications, in hundreds of cell types [4]. Common usage today defines epigenetics as chromatin-based mechanisms that can alter gene expression without changes to the DNA sequence per se [5•,6,7•] (Fig. 1).
Figure 1. Chromatin-based mechanisms can regulate gene expression profiles.
Epigenetics encompasses three nuclear processes: (1) DNA methylation, (2) histone density and posttranslational modifications, and (3) RNA-based mechanisms. DNA methylation occurs symmetrically at CpG dinucleotides, and is responsible for gene silencing. Recently described hydroxymethyla-tion is also present on DNA. Histone density can affect the accessibility of the chromatin to chromatin remodelers and transcription factors. Posttranslational modifications on N-terminal tails of histone proteins can modulate the interactions of histone proteins with DNA. RNA-based mechanisms include the production of long noncoding RNA (lncRNA), which can interact with chromatin and chromatin-modifying complexes to regulate gene expression. Adapted with permission from [5•].
DNA methylation
The idea that a heritable, postreplicative modification of DNA, or DNA methylation, can function to control gene expression was first described in the 1970s and 1980s [8,9]. This key work was seminal in defining an inverse correlation between gene activity and DNA methylation, demonstrated that this postsynthetic modification of DNA could be passed on during mitosis and DNA replication, and defined that DNA methylation silences genes through changes in chromatin structure [10,11]. Although some key processes are still murky, especially DNA demethylation pathways, we know the relevance is high. Differential DNA methylation, its presence or absence, contributes fundamentally to cell differentiation, embryonic development, stem cell biology, X-chromosome inactivation and genomic imprinting processes, and cancer pathogenesis [12,13]. It is remarkable how little we know about DNA methylation in cardiovascular disease. We have hints of relevance, but we do not have wisdom.
In vertebrates, DNA methylation occurs at carbon 5 of cytosine, almost exclusively at CpG dinucleotides, and can lead to transcriptional repression. Methyl groups may sterically hinder transcription factor binding [14]. A good example is hypoxia-inducible factor (HIF), which has a CpG dinucleotide in the cis-recognition element. Other trans-factors are not affected by CpG methylation (e.g. Sp1). The family of methyl-CpG-binding proteins, including MeCP2, can block access of transcription factors to the promoter by interacting with methylated CpG sites or by recruiting other histone-modifying enzymes that repress transcription [15–19]. Genetic defects in MeCP2 result in the neurodevelopmental disorder, Rett syndrome. Proteins have been characterized that preferentially bind to unmethylated CpG dinucleotides, which may have important functions in preserving nonmethylated regions of chromatin (e.g. Cfp1, a cysteine-rich CXXC domain protein) [20,21••]. Further characterization of the proteins that interpret CpG marks will aid in elucidating how chromatin modifications can regulate gene expression.
Mammals have three DNA methyltransferases (DNMTs), namely DNMT1, DNMT3a, and DNMT3b, which catalyze the addition of a methyl moiety specifically at CpG dinucleotides [13]. DNMT1 is regarded as the maintenance methyltransferase. DNMT1 permits conservative transmission of an epigenetic mark via remethylation of a hemimethylated nascent DNA strand. It is interesting that the fidelity of this process is very high. Yet, and perhaps important in disease, compared with conservation of A, C, G, T fidelity with DNA replication, DNA methylation of methyl-cytosine is log orders less efficient. DNMT3a and 3b are de-novo methyltransferases and play crucial roles in embryonic development [14]. These enzymes can set down new patterns of DNA methylation on unmethylated DNA.
Two processes of DNA demethylation have been described – passive (replication independent) and active (replication dependent) pathways [22]. Interestingly, the recent discovery that 5-methylcytosine can be oxidized to 5-hydroxymethylcytosine in mammals by the TET1, TET2, and TET3 enzymes has suggested intriguing possibilities, especially as the biological role of this modification remains unknown [23,24]. Some argue that the production of 5-hydroxymethylcytosine may be an intermediate in DNA demethylation pathways, highlighting an important area of focus for future studies.
Histone posttranslational modifications
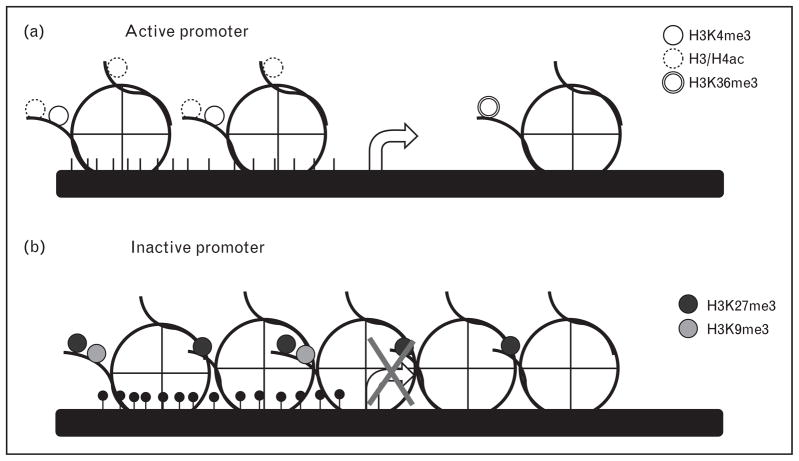
Chromatin is compacted DNA wound around histone and nonhistone proteins. The nucleosome serves as the fundamental repeating unit, composed of an octamer of two copies of each of the histone proteins H2A, H2B, H3, and H4. Numerous covalent modifications are possible, especially on the N-termini of histone tails [25]. Two of the best known marks include lysine acetylation and lysine methylation [26] (Table 1). Hyperacetylated histones H3 and H4 are found at the promoter regions of genes and are correlated with transcriptional activation. In contrast, trimethylation of lysine 27 or lysine 9 on histone H3 has been shown to result in gene silencing [25] (Fig. 2). One well characterized chromatin domain consists of trimethylation of histone H3 lysine 4 at the promoter of the gene and trimethylation of histone H3 lysine 36 on the body of the gene, commonly referred to as the K4–K36 chromatin domain, which, along with RNA polymerase II binding, defines transcriptionally active regions of the genome [27]. Recently, these K4–K36 domains have been helpful in functionally characterizing chromatin, and have been especially useful in identifying large numbers of previously uncharacterized promoters for genes [28,29]. Important areas to follow are the relevance of these paradigms for genes that are active in cells of the cardiovascular lineage and the relative importance of these transcriptional control pathways as mediators of disease.
Table 1.
A summary of histone posttranslational modifications and their effects on gene expression
| Histone modification | Location | Effect on gene expression |
|---|---|---|
| Histone H3 | ||
| K9ac | Promoter | On |
| K14ac | Promoter | On |
| K4me3 | Promoter | On |
| K9me3 | Promoter, heterochromatin | Off |
| K27me3 | Promoter, heterochromatin | Off |
| K36me3 | Transcribed region | On |
| S10phos | Promoter | On/off |
| T6phos | Promoter | Off |
| T11phos | Promoter | Off |
| Histone H4 | ||
| K5ac | Promoter | On |
| K8ac | Promoter | On |
| K12ac | Promoter | On |
| K16ac | Promoter | On |
| K20me3 | Heterochromatin | Off |
| Histone H2A | ||
| K119ub | Promoter | Off |
| Histone H2B | ||
| K120ub | Unknown | On |
ac, acetylation; K, lysine; me3, trimethylation; phos, phosphorylation; S, serine; T, threonine; ub, ubiquitination.
Figure 2. Signature chromatin domains at active and inactive gene promoters.
Specific chromatin marks have been characterized at active (a) and inactive (b) genes, where the arrow denotes the start of transcription. (a) Generally, activated promoters are characterized by an absence of promoter DNA methylation (CpG sites shown as black lines), an enrichment of hyperacetylated histones H3 and H4, H3K4me3 at the promoter and H3K36me3 along the transcribed region. Transcribed regions do not demonstrate H3K27me3 marks. (b) Inactive promoters are generally characterized by dense DNA methylation (shown as filled circles at CG sites), increased histone density, and enrichment of H3K27me3 and/or H3K9me3 marks.
Two families of proteins that mediate the addition and removal of acetylated lysines are histone acetyltransferases (HATs) and histone deacetylases (HDACs) [30]. HDACs are organized into four classes of proteins, based on their homology to yeast HDACs: class I (HDAC1-3, HDAC8), class II (HDAC4-7, HDAC9-10), class III (SIRT1-7), and class IV (HDAC11) [25]. Tri-chostatin A (TSA) is a pharmacological inhibitor of class I and II HDACs and, among other approaches, can be used to address the functional relevance of histone acetylation with respect to gene expression.
RNA-based mechanisms
A newer area of epigenetics encompasses RNA-based mechanisms, which include gene regulation through long noncoding RNAs (lncRNAs). These are functionally distinct from small noncoding RNAs, such as micro-RNAs. One of the most studied lncRNAs is the 17 kb Xist nuclear RNA, which is expressed exclusively from the inactivated X-chromosome (Xi) in women, and is essential for its silencing in XX female cells [31]. Also, exciting findings using K4–K36 chromatin domain signatures to demarcate transcriptional units have identified the existence of thousands of lncRNAs in mammalian cells with broad cellular functions [29]. Since the initial reports in 2008 and 2009, emphasis has focused on defining their functional interactions with chromatin-modifying complexes [32]. One such lncRNA, HOTAIR, was found to regulate the expression of developmental HOX genes and has since been implicated in increasing the invasiveness and metastasis of breast cancer [33,34••].
Epigenetic regulation of vascular endothelium genes
We and others have demonstrated a loss of endothelial nitric-oxide synthase (eNOS) expression in human endothelial cells overlying advanced atherosclerotic lesions [35,36]. Significantly, decreased eNOS mRNA and protein levels are observed in endothelial cells overlying the neointimal lesion [35]. In contrast, we found increased expression of all three nitric-oxide synthase (NOS) isoforms (eNOS, iNOS, and nNOS) in the atherosclerotic neointima, including increased eNOS mRNA expression in neointimal macrophages and vascular smooth muscle cells [35]. How can eNOS be repressed in endothelial cells, where it is normally expressed, while in the same tissues one can observe loss of the gene repression of eNOS in VSMC, where it is not normally expressed?
Investigations into the transcriptional processes regulating two nitric oxide synthase enzymes, eNOS and inducible nitric-oxide synthase (iNOS), have elucidated classical cis/trans mechanisms of control. The significance of chromatin-based mechanisms in the transcriptional regulation of eNOS became apparent in a series of transient transfection experiments. eNOS promoter–reporter insertional transgenes are restricted in expression to endothelial cells in the murine setting [37–39]. Surprisingly, transfected episomes displayed robust expression, regardless of cell type, in culture [40]. We demonstrated an epigenetic basis for this differential expression by identifying hypomethylated CpGs in the eNOS promoter of eNOS-expressing endothelial cells, in stark contrast to dense DNA methylation in non-eNOS-expressing cells (e.g. VSMC) [40]. Further studies have interrogated the histone modifications present at the eNOS proximal promoter, and found an enrichment of acetylated histones H3 and H4 and methylated lysine 4 of histone H3, modifications associated with actively transcribed chromatin in endothelial cells [41]. The functional relevance of these pathways was addressed. Treatment of VSMC with inhibitors of DNA methyltransferase activity, such as 5-azacytidine, led to increases in eNOS mRNA in these cells. Furthermore, treatment of VSMC with inhibitors of HDAC activity, such as trichostatin A, led to increases in eNOS mRNA in these cells and increased H3 and H4 acetylation at the eNOS proximal promoter. Taken together, these studies indicated that several epigenetic processes work in concert to regulate the endothelial cell-restricted expression of eNOS. An important conceptual realization is that the eNOS gene is being actively silenced in VSMC by repressive epigenetic marks. Not surprisingly, chromatin immunoprecipitation assays indicate that MeCP2 and HDAC-1 and HDAC-2 are basally engaged at the eNOS promoter in VSMC. The reason this concept is important is that it represents a paradigm shift. Most studies in the cardiovascular system have emphasized the role of classical cis/trans pathways in the transcriptional activation of cell-restricted genes in the cell type in which they are expressed. These epigenetic studies now focus attention on cell types in which the genes are not expressed.
The inducibility of iNOS in response to cytokine stimulation in cultured human cells is highly cell-type specific [42–45]. In cultured human endothelial cells, in which iNOS is noninducible, dense methylation of the iNOS proximal promoter has been noted [46]. This was in contrast to iNOS-inducible cultured human cell types, where light methylation was observed. In addition, we showed dimethylation and trimethylation of H3K9 at the iNOS proximal promoter in noninducible cell types, providing further evidence for cell-type specific epigenetic regulation. Therefore, the transcriptional hypo-responsiveness of the iNOS gene in human umbilical vein endothelial cells (HUVECs) was mediated, in part, via epigenetic pathways. As iNOS is expressed in endothelial cells in atherosclerotic lesions, it raises the intriguing possibility that alterations in epigenetic pathways are operative in this gene activation.
There is newer evidence that altered epigenetic states are involved in atherogenesis. Global and gene-specific changes in DNA methylation are associated with disease states, especially in cancer [47,48•]. Early studies have noted aberrant DNA methylation patterns in the atherosclerotic ApoE-null mouse model, in which it was found that DNA methylation patterns were abnormal even prior to the formation of macrovascular lesions [49]. Although these findings are preliminary, the observed changes in DNA methylation (hypermethylation and hypomethylation) occurred in transcribed genomic regions [49].
Epigenetics during development
There is a need for a greater understanding of the role of epigenetics in embryonic development, as reprogramming of cells into a specific lineage for research or clinical applications will be extremely valuable. During normal development, epigenetic marks in the genome are removed and then re-established, a process known as reprogramming [50]. The differentiation of embryonic stem cells (ESCs) is accompanied by widespread changes in gene expression profiles of many genes [51]. Epigenetic mechanisms contribute to the transcriptional control of these genes, the balance of which may impact cell identity and self-renewal capability [52]. Human ESCs display a unique DNA methylation pattern [53]. The loci of lineage-specific factors are repressed by chromatin modifiers, helping to maintain a pluripotent state. Polycomb group (PcG) protein complexes, which are transcriptional repressors, turned out to be chromatin-modifying complexes. PcG proteins function in regulating genes involved in differentiation, lineage-specific genes, and embryonic development through formation of two complexes, PRC1 and PRC2 [54]. PRC2 activity is primarily associated with trimethylation activity of H3K27 [55,56]. Profiling of ESCs localized H3K27me3 marks to inactive promoter regions, along with components of the PRC2 complex [54]. Interestingly, H3K4me3 was also found at a number of inactive promoters in ESCs, particularly at genes encoding transcription factors important for lineage specification [57]. The presence of both activating and silencing marks at the same promoter region is thought to poise the gene for activation upon differentiation, and may be critical for maintaining pluripotency.
Newer studies are investigating the potential for the in-utero environment to alter epigenetic marks. Dietary protein restriction of pregnant rats can lead to hypertension and endothelial dysfunction in the offspring [58]. Remethylation of homocysteine to methionine is a key step in the synthesis of the universal methyl donor S-adenosyl methionine (SAM) and is dependent on folate. Although not fully understood, folate deficiency can lead to abnormalities in DNA methylation. A diet with folate supplementation prevents increased blood pressure and improves endothelial function [59]. Studies like this suggest that in-utero exposure to maternal atherosclerotic risk factors can prime the vasculature of the fetus to severely aggravate neointima formation in adult life. The contribution of epigenetic pathways to these environmental ‘stressors’ warrants further study.
Epigenetics and environmental cues: hypoxia and shear stress
Hypoxia has major effects on endothelial phenotype. In general, hypoxia decreases global transcriptional activity. The HIF transcription paradigm is an ancient eukaryotic response that allows cells to adapt to changes in oxygen supply or availability. Evidence suggests that epigenetic pathways may also be relevant. For example, hypoxia induces a global decrease in H3K9 acetylation in various cells as a possible consequence of HDAC upregulation [60,61]. Consistent with decreased global transcriptional activity under hypoxic conditions, increased global H3K9 methylation, a repressive histone mark, has been observed across different cells and is attributed, in part, to increased G9a histone methyltransferase expression [62]. Considering the importance of chromatin structure to the cell-specific expression of eNOS, it was anticipated that expression of the NOS3 gene in disease states might involve changes to chromatin structure. Hypoxia decreases expression of eNOS, in part, via transcriptional repression. Hypoxia caused a rapid and sustained decrease in H3/H4 acetylation of eNOS proximal promoter histones [63••]. Surprisingly, this was mediated via histone eviction from the eNOS proximal promoter during hypoxia. This was followed by the subsequent reincorporation of histones that lacked H3/H4 acetylation. The fact that histone density, as well as histone posttranslational modifications, can be dynamically regulated by cellular oxygen content is highly relevant to diseases of the cardiovascular system. Little is known about whether DNA methylation levels are altered at specific genes under hypoxic conditions to regulate transcription.
The vascular endothelium is constantly exposed to the physical forces of circulation (especially shear stress), which can regulate the expression patterns of a unique set of genes [64,65]. This is partly attributed to trans-factor binding to the shear stress response element (SSRE), a cis element found in promoter or enhancer regions of shear stress-regulated genes like KLF2, VEGFR2, and eNOS/NOS3, among others [66]. It has also been demonstrated that chromatin-based mechanisms contribute to the transcriptional regulation of a number of these genes. In cultured HUVECs exposed to flow, an increase in global acetylation of histones H3 and H4, as well as phosphorylation of serine 10 on histone H3 (H3S10), was observed. In addition, the formation of a chromatin-remodeling complex capable of HAT activity was detected [67]. It is very interesting that laminar shear stress can recruit HAT activity in, or near, the SSRE in the human eNOS gene in vascular endothelial cells [68]. In-vitro studies exhibited robust upregulation of eNOS expression in response to laminar flow, as measured by increased Pol II binding and rate of transcription of eNOS [69]. As laminar flow can affect gene regulation via epigenetic pathways, disturbed flow may impinge on them to regulate gene expression. Whether epigenetic pathways contribute to the susceptibility of different regions in the vasculature to atherosclerosis is worth considering. The merits of considering epigenetic pathways, models and paradigms warrants further discourse.
Conclusion
The role of epigenetic pathways in controlling gene expression represents a fundamentally new perspective on human cardiovascular disease. That epigenetic mechanisms are highly dynamic processes that can adapt and respond to environmental cues, like hemodynamic forces or hypoxia, is a newer paradigm. This perspective is especially exciting given the potential for therapeutic intervention and reprogramming of cells at the epigenetic level.
Key points.
Epigenetic mechanisms, specifically DNA methylation and specific histone modifications, are important in regulating the expression of genes enriched within the vascular endothelium.
Epigenetic processes are dynamic processes that respond to cues such as the physical forces of circulation and hypoxia.
Epigenome mapping can help define relationships between our genes and the environment.
Acknowledgments
P.A.M. is a recipient of a Career Investigator Award from the Heart and Stroke Foundation of Canada and is supported by a grant from the Canadian Institutes of Health Research (CIHR MOP 79475). A.V.S. is a recipient of the CIHR Collaborative Graduate Training Program in Molecular Medicine award. We gratefully thank members of the Marsden lab for critical review of this manuscript.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Additional references related to this topic can also be found in the Current World Literature section in this issue (pp. 271–272).
- 1•.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363:166–176. doi: 10.1056/NEJMra0905980. This overview looks at the applications and limitations of GWASs and how to use them appropriately to identify single-nucleotide polymorphisms related to disease. [DOI] [PubMed] [Google Scholar]
- 2•.Musunuru K, Strong A, Frank-Kamenetsky M, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. This study identified a locus strongly associated with both plasma low-density lipoprotein cholesterol and myocardial infarction in humans using GWAS, and also demonstrated that common noncoding DNA variants identified by GWASs can directly contribute to clinical phenotypes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3•.Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465:721–727. doi: 10.1038/nature09230. This recent overview provides an introduction to epigenetic factors and mechanisms, and how they react to environmental cues. [DOI] [PubMed] [Google Scholar]
- 4.Abbott A. Project set to map marks on genome. Nature. 2010;463:596–597. [PubMed] [Google Scholar]
- 5•.Yan MS, Matouk CC, Marsden PA. Epigenetics of the vascular endothelium. J Appl Physiol. 2010;109:916–926. doi: 10.1152/japplphysiol.00131.2010. An overview of epigenetic mechanisms important in the regulation of genes expressed in the vascular endothelium. [DOI] [PubMed] [Google Scholar]
- 6.Matouk CC, Marsden PA. Epigenetic regulation of vascular endothelial gene expression. Circ Res. 2008;102:873–887. doi: 10.1161/CIRCRESAHA.107.171025. [DOI] [PubMed] [Google Scholar]
- 7•.Bruneau BG. Epigenetic regulation of the cardiovascular system: introduction to a review series. Circ Res. 2010;107:324–326. doi: 10.1161/RES.0b013e3181f17dfe. This introduction to a review series highlights key advances in epigenetic and chromatin mechanisms involved in the regulation of the cardiovascular system. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pollack Y, Stein R, Razin A, Cedar H. Methylation of foreign DNA sequences in eukaryotic cells. Proc Natl Acad Sci U S A. 1980;77:6463–6467. doi: 10.1073/pnas.77.11.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stein R, Gruenbaum Y, Pollack Y, et al. Clonal inheritance of the pattern of DNA methylation in mouse cells. Proc Natl Acad Sci U S A. 1982;79:61–65. doi: 10.1073/pnas.79.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naveh-Many T, Cedar H. Active gene sequences are undermethylated. Proc Natl Acad Sci U S A. 1981;78:4246–4250. doi: 10.1073/pnas.78.7.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keshet I, Lieman-Hurwitz J, Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986;44:535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- 12.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 13.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 14.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Meehan RR, Lewis JD, McKay S, et al. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 16.Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64:1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- 17.Lewis JD, Meehan RR, Henzel WJ, et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 18.Nan X, Ng HH, Johnson CA, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 19.Agarwal N, Hardt T, Brero A, et al. MeCP2 interacts with HP1 and modulates its heterochromatin association during myogenic differentiation. Nucleic Acids Res. 2007;35:5402–5408. doi: 10.1093/nar/gkm599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Voo KS, Carlone DL, Jacobsen BM, et al. Cloning of a mammalian transcriptional activator that binds unmethylated CpG motifs and shares a CXXC domain with DNA methyltransferase, human trithorax, and methyl-CpG binding domain protein 1. Mol Cell Biol. 2000;20:2108–2121. doi: 10.1128/mcb.20.6.2108-2121.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21••.Thomson JP, Skene PJ, Selfridge J, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. This important study demonstrated that the protein Cfp1 preferentially binds nonmethylated CpG islands, and regions also associated with H3K4me3 peaks. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- 23.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer. Part I: Covalent histone modifications. Trends Mol Med. 2007;13:363–372. doi: 10.1016/j.molmed.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 26.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guenther MG, Levine SS, Boyer LA, et al. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mikkelsen TS, Ku M, Jaffe DB, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large noncoding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 31.Erwin JA, Lee JT. New twists in X-chromosome inactivation. Curr Opin Cell Biol. 2008;20:349–355. doi: 10.1016/j.ceb.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic non-coding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34••.Gupta RA, Shah N, Wang KC, et al. Long noncoding RNA HOTAIR repro-grams chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. This article describes the first finding of aberrant lncRNA expression in the HOX loci in breast cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilcox JN, Subramanian RR, Sundell CL, et al. Expression of multiple isoforms of nitric oxide synthase in normal and atherosclerotic vessels. Arterioscler Thromb Vasc Biol. 1997;17:2479–2488. doi: 10.1161/01.atv.17.11.2479. [DOI] [PubMed] [Google Scholar]
- 36.Oemar BS, Tschudi MR, Godoy N, et al. Reduced endothelial nitric oxide synthase expression and production in human atherosclerosis. Circulation. 1998;97:2494–2498. doi: 10.1161/01.cir.97.25.2494. [DOI] [PubMed] [Google Scholar]
- 37.Teichert AM, Miller TL, Tai SC, et al. In vivo expression profile of an endothelial nitric oxide synthase promoter-reporter transgene. Am J Physiol Heart Circ Physiol. 2000;278:H1352–H1361. doi: 10.1152/ajpheart.2000.278.4.H1352. [DOI] [PubMed] [Google Scholar]
- 38.Guillot PV, Liu L, Kuivenhoven JA, et al. Targeting of human eNOS promoter to the Hprt locus of mice leads to tissue-restricted transgene expression. Physiol Genomics. 2000;2:77–83. doi: 10.1152/physiolgenomics.2000.2.2.77. [DOI] [PubMed] [Google Scholar]
- 39.Teichert AM, Scott JA, Robb GB, et al. Endothelial nitric oxide synthase gene expression during murine embryogenesis: commencement of expression in the embryo occurs with the establishment of a unidirectional circulatory system. Circ Res. 2008;103:24–33. doi: 10.1161/CIRCRESAHA.107.168567. [DOI] [PubMed] [Google Scholar]
- 40.Chan Y, Fish JE, D’Abreo C, et al. The cell-specific expression of endothelial nitric-oxide synthase: a role for DNA methylation. J Biol Chem. 2004;279:35087–35100. doi: 10.1074/jbc.M405063200. [DOI] [PubMed] [Google Scholar]
- 41.Fish JE, Matouk CC, Rachlis A, et al. The expression of endothelial nitric-oxide synthase is controlled by a cell-specific histone code. J Biol Chem. 2005;280:24824–24838. doi: 10.1074/jbc.M502115200. [DOI] [PubMed] [Google Scholar]
- 42.Nussler AK, Di Silvio M, Liu ZZ, et al. Further characterization and comparison of inducible nitric oxide synthase in mouse, rat, and human hepatocytes. Hepatology. 1995;21:1552–1560. [PubMed] [Google Scholar]
- 43.Orpana A, Ranta V, Mikkola T, et al. Inducible nitric oxide and prostacyclin productions are differently controlled by extracellular matrix and cell density in human vascular endothelial cells. J Cell Biochem. 1997;64:538–546. [PubMed] [Google Scholar]
- 44.Rosenkranz-Weiss P, Sessa WC, Milstien S, et al. Regulation of nitric oxide synthesis by proinflammatory cytokines in human umbilical vein endothelial cells. Elevations in tetrahydrobiopterin levels enhance endothelial nitric oxide synthase specific activity. J Clin Invest. 1994;93:2236–2243. doi: 10.1172/JCI117221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, Laubach VE, Alley EW, et al. Transcriptional basis for hyporesponsiveness of the human inducible nitric oxide synthase gene to lipopolysaccharide/interferon-gamma. J Leukoc Biol. 1996;59:575–585. doi: 10.1002/jlb.59.4.575. [DOI] [PubMed] [Google Scholar]
- 46.Chan GC, Fish JE, Mawji IA, et al. Epigenetic basis for the transcriptional hyporesponsiveness of the human inducible nitric oxide synthase gene in vascular endothelial cells. J Immunol. 2005;175:3846–3861. doi: 10.4049/jimmunol.175.6.3846. [DOI] [PubMed] [Google Scholar]
- 47.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48•.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. This review provides an in-depth look at DNA methylation mechanisms and its associated pathways, in health and in cancer. [DOI] [PubMed] [Google Scholar]
- 49.Lund G, Andersson L, Lauria M, et al. DNA methylation polymorphisms precede any histological sign of atherosclerosis in mice lacking apolipoprotein E. J Biol Chem. 2004;279:29147–29154. doi: 10.1074/jbc.M403618200. [DOI] [PubMed] [Google Scholar]
- 50.Morgan HD, Santos F, Green K, et al. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14(Spec No 1):R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 51.Ivanova NB, Dimos JT, Schaniel C, et al. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 52.Bibikova M, Laurent LC, Ren B, et al. Unraveling epigenetic regulation in embryonic stem cells. Cell Stem Cell. 2008;2:123–134. doi: 10.1016/j.stem.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 53.Bibikova M, Chudin E, Wu B, et al. Human embryonic stem cells have a unique epigenetic signature. Genome Res. 2006;16:1075–1083. doi: 10.1101/gr.5319906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boyer LA, Plath K, Zeitlinger J, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 55.Kuzmichev A, Nishioka K, Erdjument-Bromage H, et al. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 57.Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 58.Brawley L, Itoh S, Torrens C, et al. Dietary protein restriction in pregnancy induces hypertension and vascular defects in rat male offspring. Pediatr Res. 2003;54:83–90. doi: 10.1203/01.PDR.0000065731.00639.02. [DOI] [PubMed] [Google Scholar]
- 59.Torrens C, Brawley L, Anthony FW, et al. Folate supplementation during pregnancy improves offspring cardiovascular dysfunction induced by protein restriction. Hypertension. 2006;47:982–987. doi: 10.1161/01.HYP.0000215580.43711.d1. [DOI] [PubMed] [Google Scholar]
- 60.Johnson AB, Denko N, Barton MC. Hypoxia induces a novel signature of chromatin modifications and global repression of transcription. Mutat Res. 2008;640:174–179. doi: 10.1016/j.mrfmmm.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnson AB, Barton MC. Hypoxia-induced and stress-specific changes in chromatin structure and function. Mutat Res. 2007;618:149–162. doi: 10.1016/j.mrfmmm.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen H, Yan Y, Davidson TL, et al. Hypoxic stress induces dimethylated histone H3 lysine 9 through histone methyltransferase G9a in mammalian cells. Cancer Res. 2006;66:9009–9016. doi: 10.1158/0008-5472.CAN-06-0101. [DOI] [PubMed] [Google Scholar]
- 63••.Fish JE, Yan MS, Matouk CC, et al. Hypoxic repression of endothelial nitric-oxide synthase transcription is coupled with eviction of promoter histones. J Biol Chem. 2010;285:810–826. doi: 10.1074/jbc.M109.067868. This important study demonstrates that hypoxia results in a decrease in H3/H4 acetylation of eNOS proximal promoter histones, which is mediated by histone eviction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gimbrone MA, Jr, Resnick N, Nagel T, et al. Hemodynamics, endothelial gene expression, and atherogenesis. Ann N Y Acad Sci. 1997;811:1–10. doi: 10.1111/j.1749-6632.1997.tb51983.x. discussion 10–11. [DOI] [PubMed] [Google Scholar]
- 65.Dekker RJ, van Soest S, Fontijn RD, et al. Prolonged fluid shear stress induces a distinct set of endothelial cell genes, most specifically lung Kruppel-like factor (KLF2) Blood. 2002;100:1689–1698. doi: 10.1182/blood-2002-01-0046. [DOI] [PubMed] [Google Scholar]
- 66.Khachigian LM, Anderson KR, Halnon NJ, et al. Egr-1 is activated in endothelial cells exposed to fluid shear stress and interacts with a novel shear-stress-response element in the PDGF A-chain promoter. Arterioscler Thromb Vasc Biol. 1997;17:2280–2286. doi: 10.1161/01.atv.17.10.2280. [DOI] [PubMed] [Google Scholar]
- 67.Illi B, Nanni S, Scopece A, et al. Shear stress-mediated chromatin remodeling provides molecular basis for flow-dependent regulation of gene expression. Circ Res. 2003;93:155–161. doi: 10.1161/01.RES.0000080933.82105.29. [DOI] [PubMed] [Google Scholar]
- 68.Chen W, Bacanamwo M, Harrison DG. Activation of p300 histone acetyl-transferase activity is an early endothelial response to laminar shear stress and is essential for stimulation of endothelial nitric-oxide synthase mRNA transcription. J Biol Chem. 2008;283:16293–16298. doi: 10.1074/jbc.M801803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Won D, Zhu SN, Chen M, et al. Relative reduction of endothelial nitric-oxide synthase expression and transcription in atherosclerosis-prone regions of the mouse aorta and in an in vitro model of disturbed flow. Am J Pathol. 2007;171:1691–1704. doi: 10.2353/ajpath.2007.060860. [DOI] [PMC free article] [PubMed] [Google Scholar]