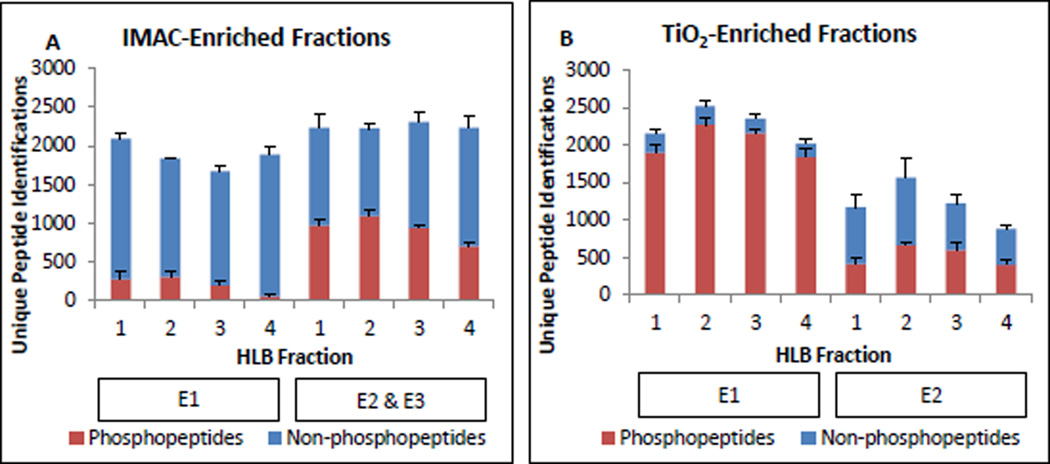
Figure 2.
IMAC-HLB and TiO2-HLB workflow performance. All values shown are the average number of unique sequences identified per biological replicate and error bars display standard deviation. Phosphosite localization and SILAC quantification status were not considered at this stage. A. Unique peptide identifications by IMAC enrichment step (E1, E2&3) and HLB fraction number. Phosphopeptides are shown in red bars; non-phosphorylated peptides are in blue. B. TiO2-enriched peptide identifications by enrichment step (E1, E2) and HLB fraction number.