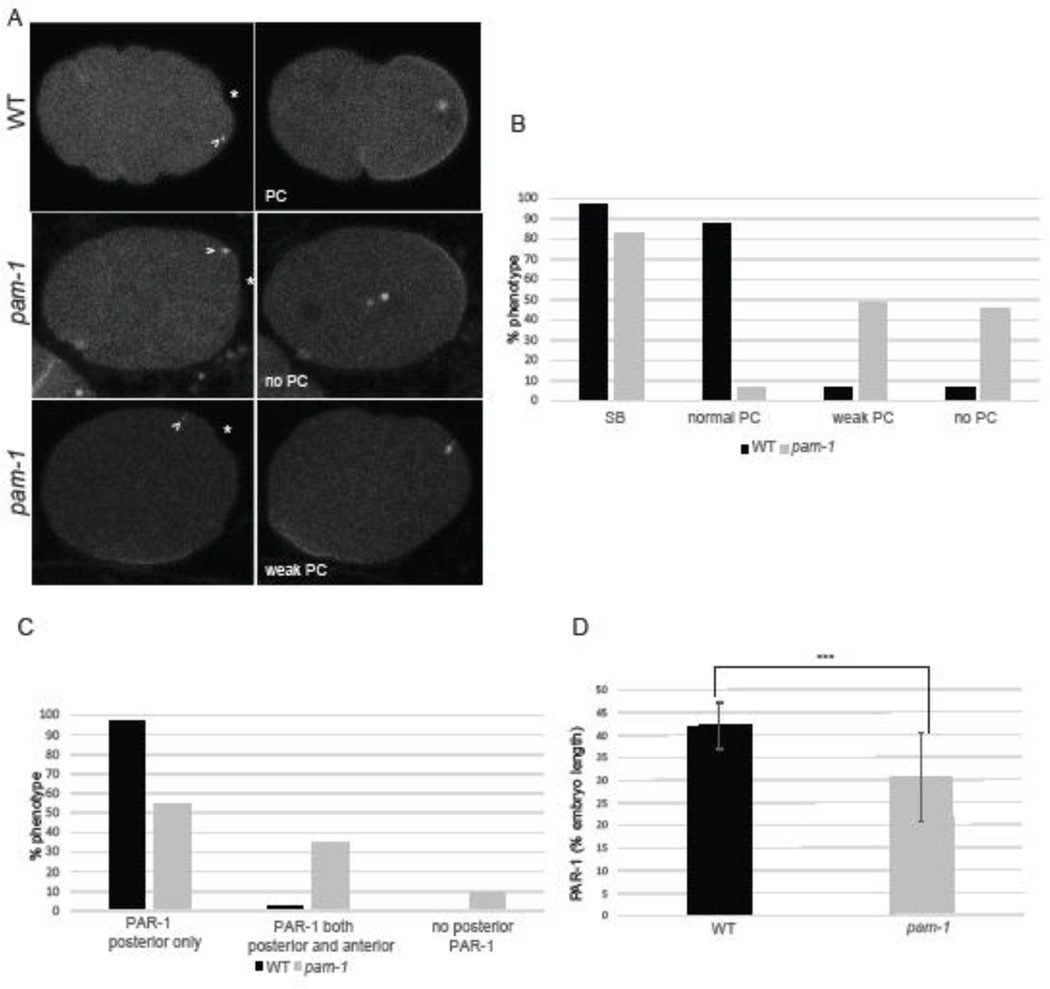
Figure 1. pam-1 mutant embryos show defects in the establishment of polarity.
A) In wild-type embryos, the centrosome (arrowhead; γ-tub::GFP) appears close to the posterior cortex and triggers symmetry breaking (SB) (*). This is followed by pseudocleavage (PC) and localization of PAR-1 (GFP) to the posterior cortex. In pam-1 mutants, the centrosome is positioned similarly to wild-type at first and most embryos exhibit SB. However, most embryos exhibit no or weak PC and small or mislocalized PAR-1 domains. Posterior is to the right in all images B) The percentage of wild-type and pam-1 mutant embryos with different polarity phenotypes (WT=32; pam-1 =64). C) The percentage of wild-type (n=33) and pam-1 mutant (n=51) embryos with different PAR-1 localizations, as documented at the time of pronuclear envelope breakdown. D) pam-1 mutant embryos that localize PAR-1 posteriorly (n=46) exhibit smaller domains as compared to wild-type (n=32). Standard deviation is shown. *** p < 0.0001.