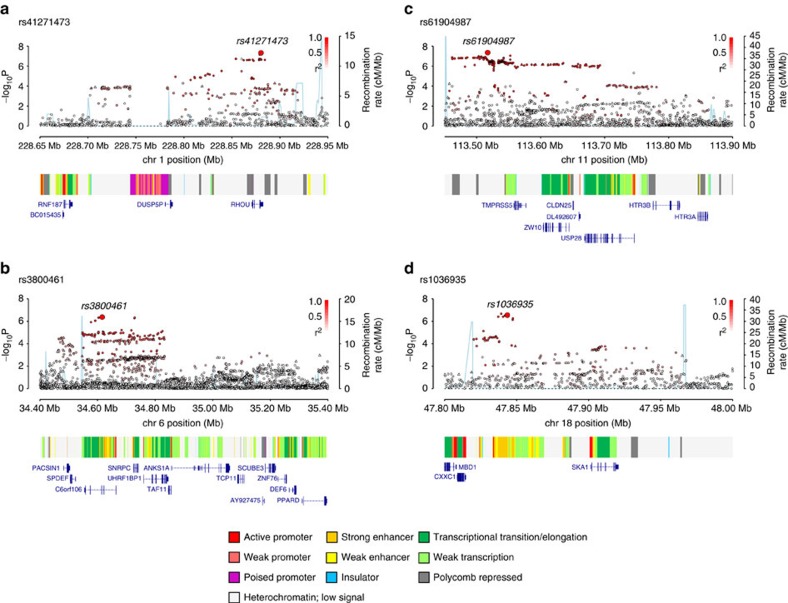
Figure 3. Regional plots of association results and recombination rates for new risk loci for chronic lymphocytic leukaemia.
Results shown for 1q42.13, 6p21.31, 11q23.2, 18q21.1 (a–d). Plots (drawn using visPig62) show association results of both genotyped (triangles) and imputed (circles) SNPs in the GWAS samples and recombination rates. −log10 P values (y axes) of the SNPs are shown according to their chromosomal positions (x axes). The sentinel SNP in each combined analysis is shown as a large circle or triangle and is labelled by its rsID. The colour intensity of each symbol reflects the extent of LD with the top genotyped SNP, white (r2=0) through dark red (r2=1.0). Genetic recombination rates, estimated using the 1000 Genomes Project samples, are shown with a light blue line. Physical positions are based on NCBI build 37 of the human genome. Also shown are the chromatin-state segmentation track (ChromHMM) for lymphoblastoid cells using data from the HapMap ENCODE Project, and the positions of genes and transcripts mapping to the region of association.