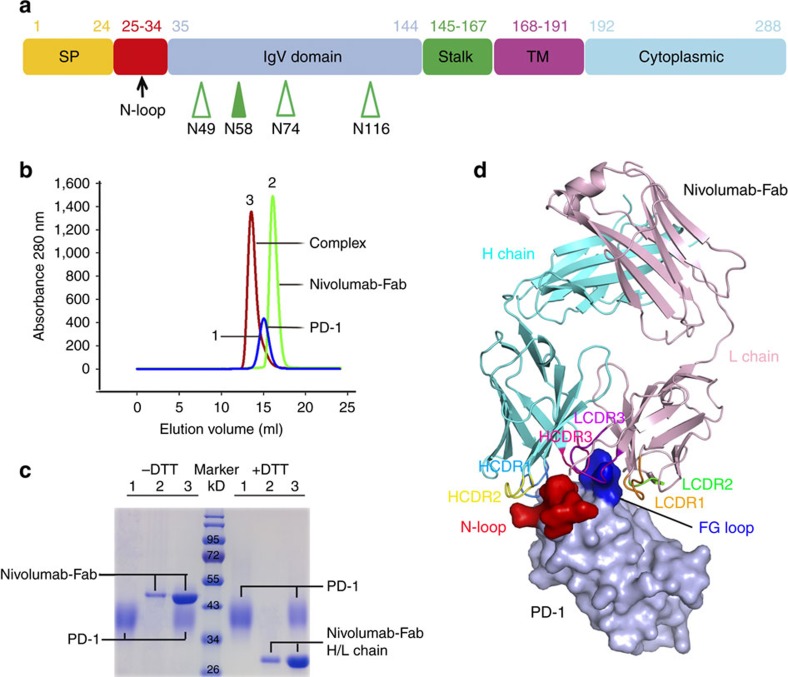
Figure 1. Overall structure of PD-1 nivolumab-Fab complex.
(a) Schematic diagram of PD-1 protein. Full-length PD-1 is separated into ectodomain, transmembrane domain and intracellular domain. Ectodomain is composed of the signal peptide, N-loop, IgV domain and stalk region. The N-glycosylation at N58, which could be seen in the complex structure in the ectodomain is indicated with solid green arrowhead, whereas the other three N-glycosylation sites are indicated with hollow green arrowheads. The numbers indicate amino acid positions. (b) Gel filtration profiles of PD-1 (blue), nivolumab-Fab (green) and the PD-1-nivolumab-Fab complex (red) were analysed by size-exclusion chromatography as indicated. (c) The separation profiles of each pooled samples on SDS-PAGE are shown in reducing (+DTT) or non-reducing (-DTT) conditions, 1 for PD-1, 2 for nivolumab-Fab and 3 for PD-1-nivolumab-Fab complex. (d) The complex structure of nivolumab-Fab bound to PD-1. The Fab fragment of nivolumab is shown as cartoon (Heavy chain, cyan; Light chain, pink), and PD-1 is shown as surface representation (light blue). The CDR1, CDR2 and CDR3 loops of the heavy chain (HCDR1, HCDR2, HCDR3) are coloured in marine, yellow and magenta, respectively. The CDR1, CDR2 and CDR3 loops of light chain (LCDR1, LCDR2, LCDR3) are coloured in orange, violet and green, respectively. The N-loop and FG loop of the PD-1 molecule are highlighted in red and blue, respectively.