Figure 2.
Mutagenesis of Conserved Amino Acids at the CENPC-k Motif as Well as Its Complete Deletion Results in KNL2 Mislocalization That Can Be Restored by the CENPC Motif.
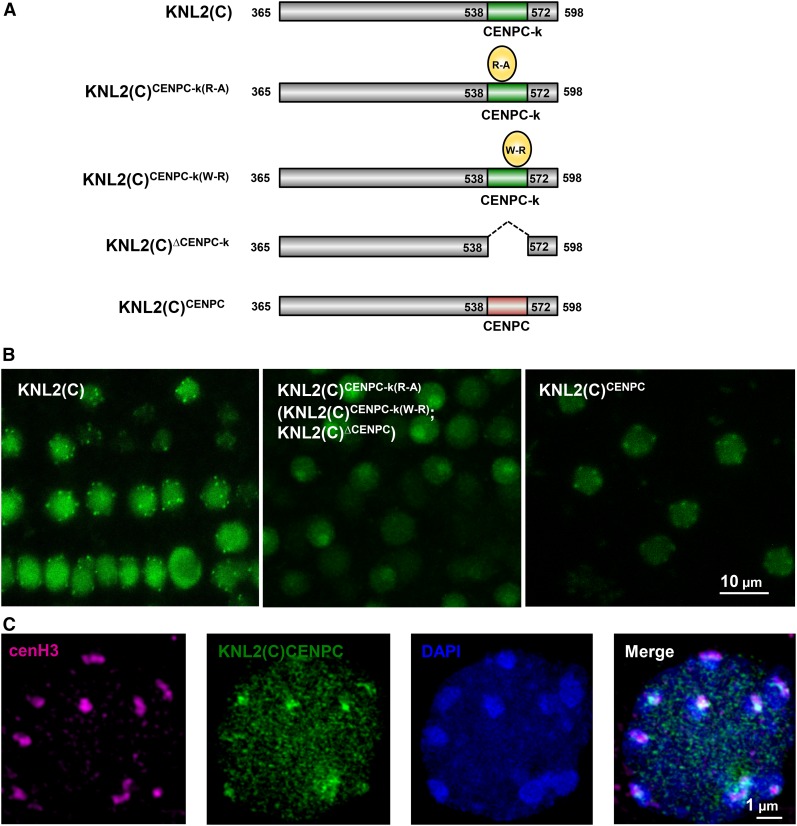
(A) Schematic view of EYFP-tagged KNL2 constructs: KNL2(C), KNL2(C)CENPC-k(R-A), KNL2(C)CENPC-k(W-R), KNL2(C)∆CENPC-k, and KNL2(C)CENPC.
(B) Subnuclear localization of KNL2(C) (left panel), KNL2(C)CENPC-k(R-A) (middle panel), and KNL2(C)CENPC (right panel) fused to EYFP in root tip nuclei of Arabidopsis transformed with corresponding constructs. KNL2(C)CENPC-k(W-R) and KNL2(C)∆CENPC-k proteins (indicated in brackets in the middle panel) showed the same localization pattern as KNL2(C)CENPC-k(R-A). The C-terminal part of KNL2 fused to EYFP showed nucleoplasmic and centromeric localization (Lermontova et al., 2013), while protein variants carrying the point mutations within the CENPC-k motif or lacking this motif completely lost an ability to localize at centromeres and were detected only at nucleoplasm and nucleolus. However, the C terminus of KNL2 with the substitution of CENPC-k by the CENPC motif was targeted to centromeres.
(C) Double immunostaining with anti-GFP [detecting the KNL2(C)CENPC EYFP fusion] and anti-cenH3 antibodies showed colocalization of both immunosignals at bright 4′,6-diamidino-2-phenylindole (DAPI)-stained chromocenters.