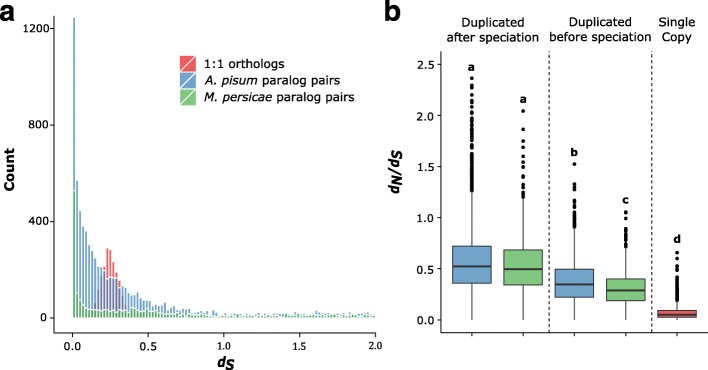
Fig. 2.
M. persicae experienced greater gene loss rates (a) and stronger purifying selection in retained ancestral duplicates (b) than A. pisum. a Age distribution of duplicated genes in M. persicae and A. pisum. The number of synonymous substitutions per synonymous site (d S) was calculated between paralog pairs for M. persicae (green) and A. pisum (blue) using the YN00 [91] model in PAML [82]. For each duplicated gene, only the most recent paralog was compared. Pairwise d S was also calculated for 1:1 orthologs between M. persicae and A. pisum (red), the peak in which corresponds to the time of speciation between the two aphid species. After filtering, 1955 M. persicae paralog pairs, 7253 A. pisum paralog pairs and 2123 1:1 orthologs were included for comparison. Mean d S of 1:1 orthologs between A. pisum and M. persicae was 0.26. b Box plots showing median d N /d S for A. pisum and M. persicae paralog pairs that duplicated before and after speciation of the two aphid species and for 1:1 orthologs between the two species. Older duplicate genes have lower d N /d S than recently duplicated genes (since speciation) indicating stronger purifying selection in ancestral versus recent duplicates. Additionally, older duplicate genes in M. persicae have significantly lower d N /d S than in A. pisum (Mann–Whitney U = 1816258, M. persicae: 1348 paralog pairs, A. pisum: 3286 paralog pairs, p = < 0.00001) indicating stronger genome streamlining in M. persicae than in A. pisum. Box plots are shaded by species as in (a)