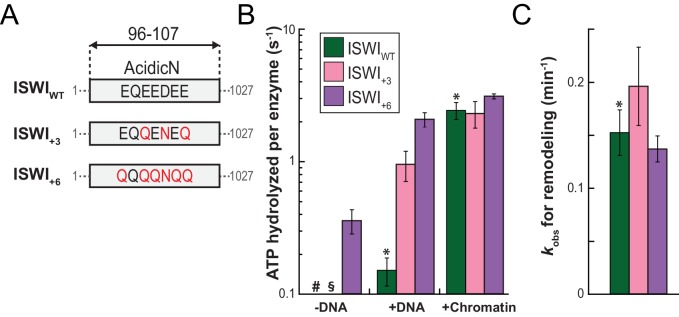
Figure 6. AcidicN is a strong negative regulator of the ATPase.
(A) Design of AcidicN derivatives of ISWI (see also Figure 6—figure supplement 1A). (B) Effects of AcidicN mutation on ATP hydrolysis in absence or presence of saturating concentrations of DNA and chromatin. In absence of DNA, ATPase activities of ISWIWT (#) and ISWI+3 (§) were ≤0.06 s−1. Errors are s.d. (n ≥ 4). (C) Effects of AcidicN mutation on the remodeling activities. Nucleosomal arrays containing wild-type H4 were used. Errors are s.d. (n ≥ 3) except for ISWI+3 for which minimal and maximal values of two independent measurements are shown. Color code as in panel (B). Raw data of the remodeling assay can be found in Figure 8—figure supplement 1. Results for ISWIWT (*) are replotted for comparison from Figure 3B,C.
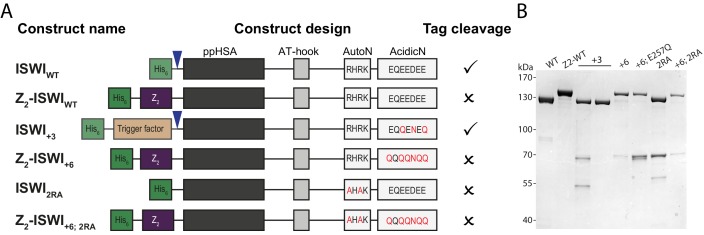
Figure 6—figure supplement 1. AcidicN and AutoN mutants.
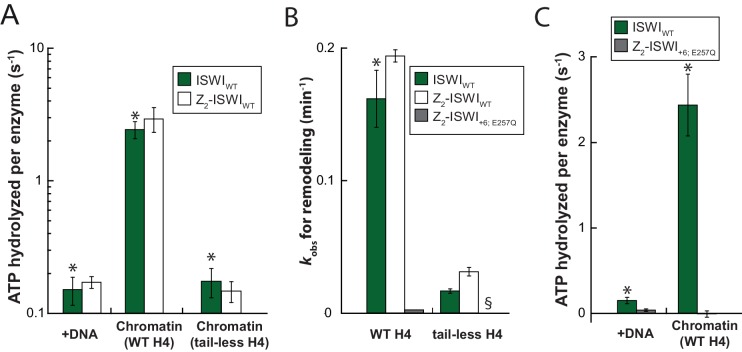
Figure 6—figure supplement 2. Comparison of ATPase and remodeling activities of ISWI control variants used in this study.
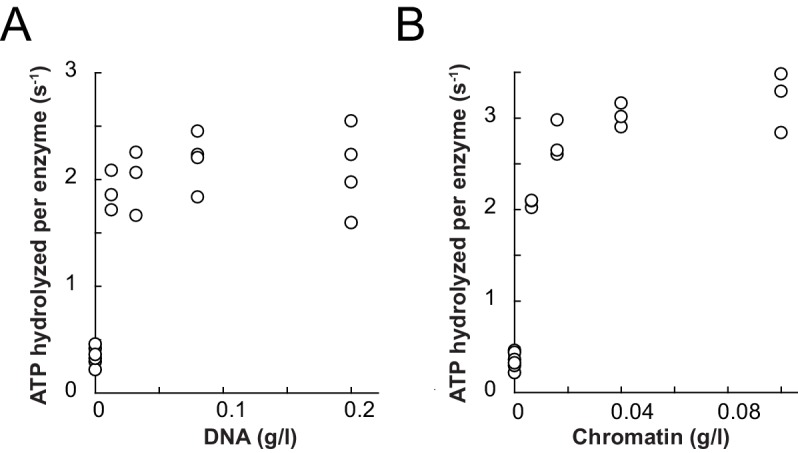
Figure 6—figure supplement 3. Saturation controls for ISWI+6 in ATPase assays.
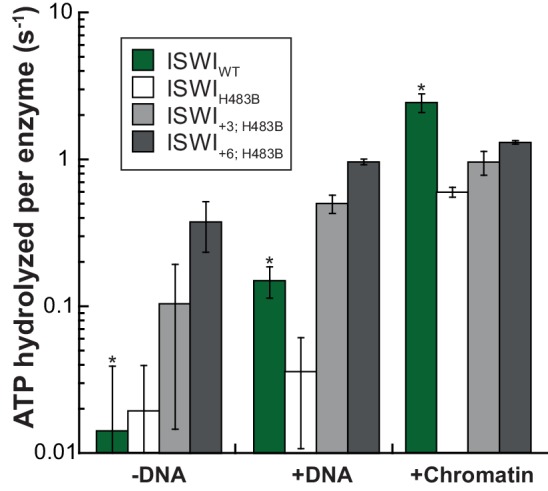
Figure 6—figure supplement 4. AcidicN mutations upregulate the ATPase activity of ISWIH483B.