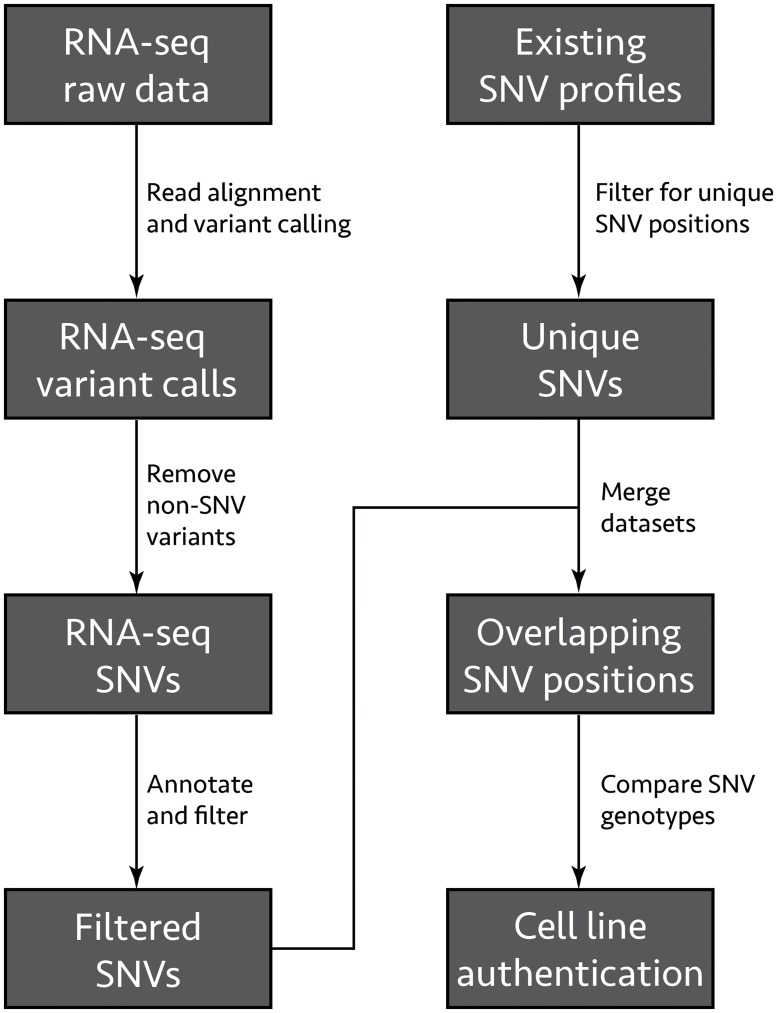
Fig 1. RNA-seq cell line authentication pipeline.
Raw RNA-seq data is aligned to the human genome using STAR, followed by processing and variant calling steps using GATK tools. Non-SNV variants (insertions, deletions, etc.) are removed, and the resulting SNVs are annotated using SnpEff and SnpSift. SNVs passing the GATK variant filtration step and having a total allelic depth of at least 10 are compared to COSMIC SNV profiles filtered to include unique SNV positions, as well as a SNV genotyping panel.