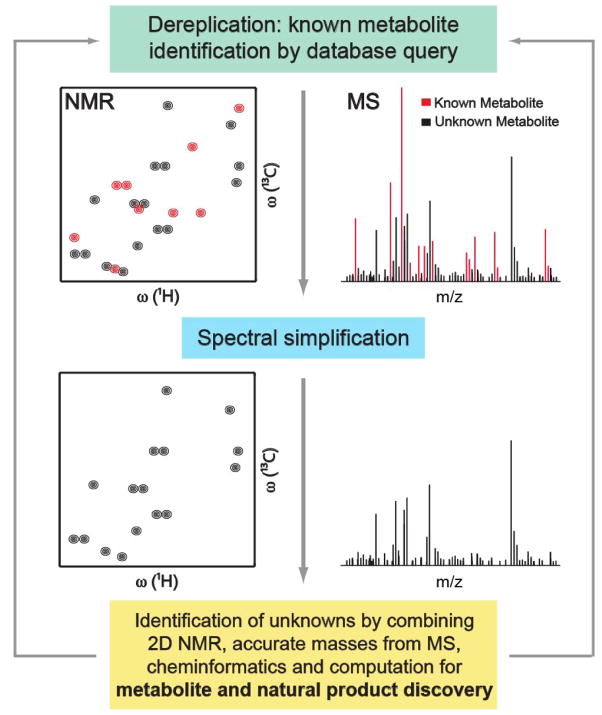
Figure 1.
Integrated metabolomics workflow for the identification of known and unknown metabolites in complex mixtures. Combined use of metabolomics databases with experimental NMR and MS spectra (e.g., NMR/MS Translator [27]) allows the rapid identification of a maximal number of known metabolites present in the mixture, while unidentified signals are used as fingerprints of unknowns. Next, structures of unknown metabolites can be elucidated or vastly narrowed down by the combined use of multidimensional NMR, MS, cheminformatics, and computation (e.g., SUMMIT MS/NMR [35]).